Figure 6.
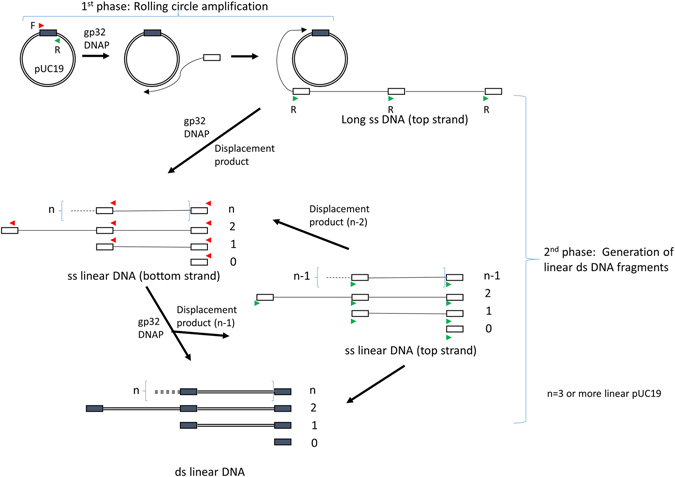
A model of gp32-assisted isothermal amplification on circular templates. The amplicon (depicted as a box) is specified by a pair of primers (forward, F, red arrow and reverse, R, green arrow) on the circular ds pUC19 template (circle). The amplification is proposed to occur in two phases. In the first phase, with the help of gp32 protein both primers (for simplicity only the product from F primer is shown) invades dsDNA and initiates DNA synthesis by a strand-displacing DNA polymerase to produce long single-stranded DNA corresponding to the top strand sequence of pUC19. For circular ssDNA template, this phase occurs similarly but does not require a strand invasion step. In the second phase, which is also facilitated by gp32, this single stranded product serves as the template for the reverse primer (R, green arrow) for DNA synthesis in the opposite direction. As there is one primer site at each amplicon for every length of the pUC19, discrete sizes of ssDNA products (bottom strand) are generated through strand displacement synthesis at multiple sites and they consist of a single amplicon and larger species of one or more entire length of the linear templates (n) linked with a single amplicon at one end. After another round of priming and DNA synthesis by the other primer (red arrows), these ssDNA species are then converted to dsDNA fragments which migrate as discrete bands in agarose gels. At the same time, ssDNA species of the opposite strand (top strand) are also generated in a similar displacement synthesis but one linear pUC19 shorter (n-1) and they are converted to ds fragments in subsequent round of synthesis.
