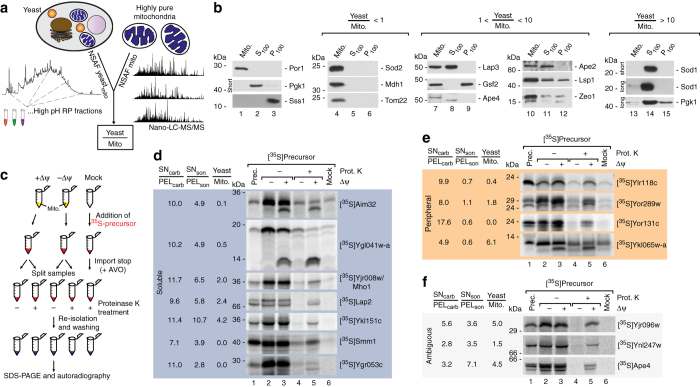
Fig. 3.
Validation of mitochondrial localization for novel proteins. a Quantitative assessment of protein abundance in total yeast cells and purified mitochondria based on spectral counting (normalized abundance factor). Resulting Yeast/Mito ratios indicate presence of proteins either exclusively in mitochondria or in multiple cellular compartments. b Immunoblot analysis of cellular fractions containing pure mitochondria (Mito), enriched cytosolic proteins (S100), or microsomal proteins (P100). Por1, mitochondrial marker; Pgk1, cytosolic marker; Sss1, ER marker. long, long exposure time; short, short exposure time. c Schematic overview of in organello import reactions to validate mitochondrial localization. [35S]labeled precursors of candidate proteins were generated by in vitro transcription/translation and incubated with isolated mitochondria in the presence or absence of the membrane potential (Δψ) or without mitochondria (Mock). Import reaction was terminated by depletion of the membrane potential and samples were treated with Proteinase K where indicated to remove non-imported precursors. Samples were analyzed by SDS–PAGE and radiolabelled proteins visualized by autoradiography. In case of presequence cleavage upon import a size shift from the precursor to the mature protein can be observed. d In organello import of indicated radiolabelled precursors of novel mitochondrial proteins which localize to the soluble protein fraction. e In organello import of indicated radiolabelled precursors of novel mitochondrial proteins which localize to the peripheral membrane protein fraction. f In organello import of protein candidates from the ambiguous fraction. Prot. K, Proteinase K; prec. precursor; Δψ, membrane potential across the inner mitochondrial membrane