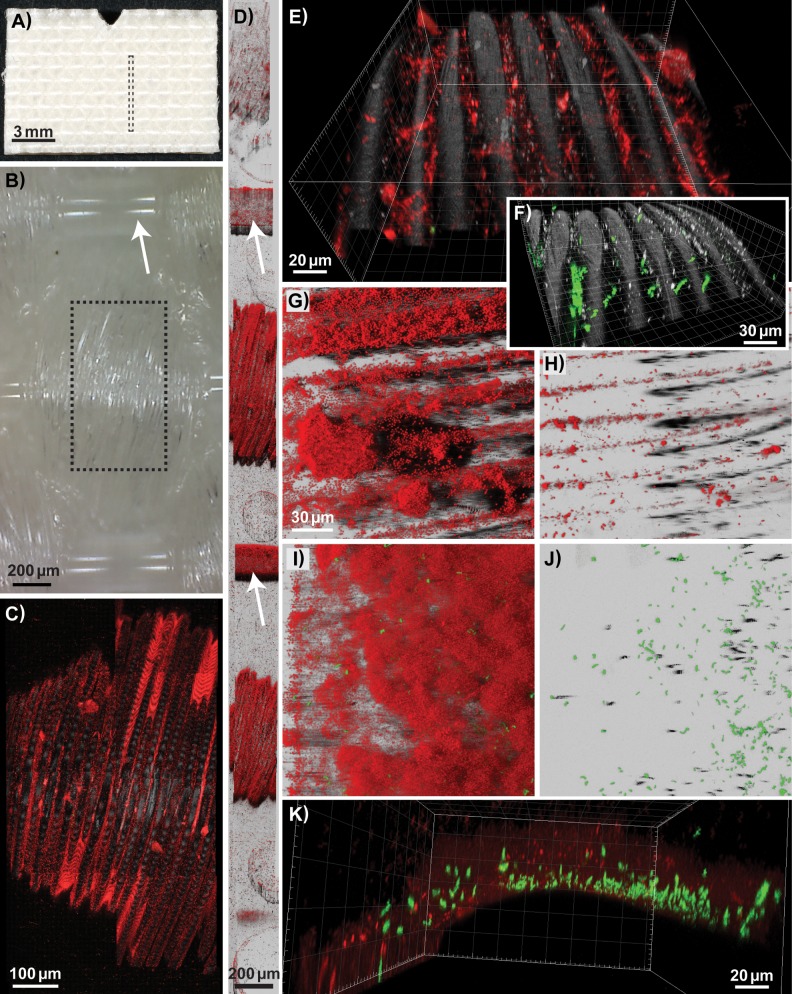
FIG 7.
Biofilms formed on the underside of the conveyor belt examined using CLSM. Pseudomonas and Acinetobacter background flora strains (shown in red) and GFP-expressing L. monocytogenes strains (in green) were used as inoculum, except in (F), where biofilms were inoculated with L. monocytogenes strains only. (A, B) Photographs showing the underside of a clean conveyor belt coupon. (B) Photograph taken with a USB microscope lens showing the weave pattern of the fabric, with the smooth warp thread indicated by an arrow. Biofilms formed on the weft thread of the woven fabric, on coupons rinsed in H2O on days 4 to 7 (C, D, and F), on a coupon rinsed in H2O on day 7 after treatment with QAC on days 4 to 6 (E), and on coupons harvested on day 4 for after treatment with PAA (G) or QAC (H). (C, E, and F) The smaller fibers constituting the weft thread are represented by the reflection signal (in gray). Mosaic 3D meta-images are shown in section view mode (C) and Easy3D blend representation (D), with warp treads indicated by arrows. The boxes drawn with dashed lines in panels A and B show the size and location of the area of a coupon depicted by the CLSM images shown in panels D and C, respectively. (I to K) Different representations of the same scan, showing a biofilm harvested on day 4 after rinsing in H2O, formed on the warp thread of the woven fabric. Images are shown as three-dimensional (E, F, and K) and Easy 3D (G, H, I, and J) IMARIS representations. The scales are the same in the Easy3D blend images in panels G to J. The red channel is not shown in panels F and J.