Figure 1.
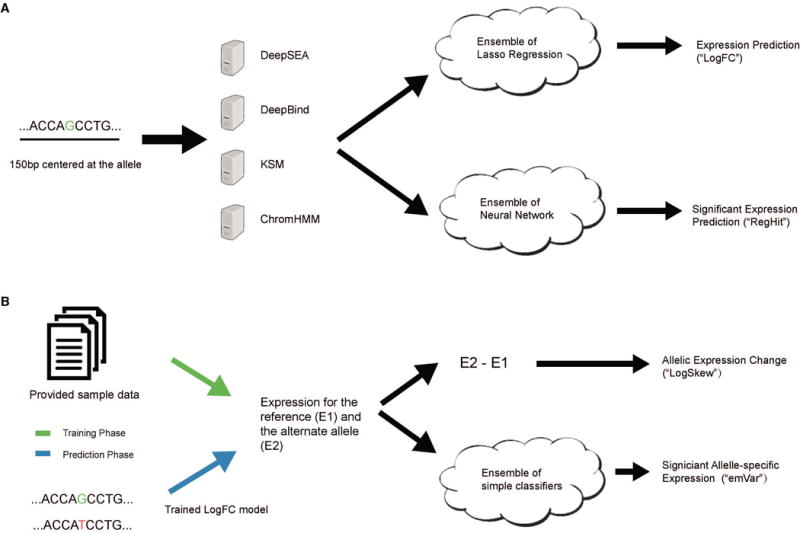
The schematic of EnsembleExpr. (A) The 150-bp sequence centered at the queried allele is taken as input to four computational models to generate functional features that are used by two ensemble models to make expression predictions (“log2FC”) and significance estimates (“RegHit”). (B) During training the provided expression levels of the two alleles of each variant are used to train an ensemble model of significant allele-specific expression (ASE). During testing we first apply the trained expression model in (A) to generate expression predictions, which are then given to the significant ASE model to make predictions (“emVar”). The difference of the predicted expression levels is directly output as the prediction for allelic expression change (“LogSkew”).
