Fig. 2.
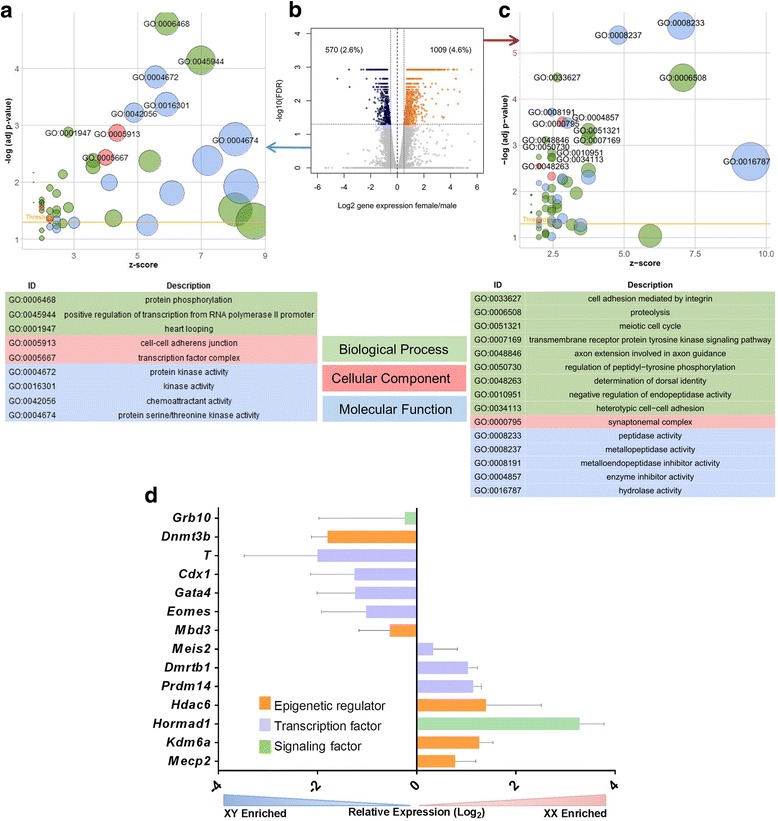
Gene ontology of differentially expressed genes between XY and XX ES cells. a, c Upregulated biological process, molecular function and cellular components between XY (a) and XX (c) ES cells. The XY lines had increased expression of genes related to early development (heart looping) and kinase activity. The XX lines showed increased expression of meiotic cell cycle-associated genes and peptidase activity. x-axis: z-score of associated terms, which indicates degree of upregulation; y-axis: significance of term. b Volcano plot showing selection of the differentially expressed genes between XY and XX lines at FDR < 0.05 (blue dots, higher in males; orange dots, higher in females). d qPCR validation of genes detected in the RNA sequencing dataset illustrating a sex-biased expression pattern of transcription factors, epigenetic enzymes and key signaling factors
