Abstract
Tenacibaculum maritimum is a devastating bacterial pathogen of wild and farmed marine fish with a broad host range and a worldwide distribution. We report here the complete genome sequence of the T. maritimum type strain NCIMB 2154T. The genome consists of a 3,435,971-base pair circular chromosome with 2,866 predicted protein-coding genes. Genes encoding the biosynthesis of exopolysaccharides, the type IX secretion system, iron uptake systems, adhesins, hemolysins, proteases, and glycoside hydrolases were identified. They are likely involved in the virulence process including immune escape, invasion, colonization, destruction of host tissues, and nutrient scavenging. Among the predicted virulence factors, type IX secretion-mediated and cell-surface exposed proteins were identified including an atypical sialidase, a sphingomyelinase and a chondroitin AC lyase which activities were demonstrated in vitro.
Keywords: Tenacibaculum maritimum, fish pathogen, virulence factors, genome, toxins
Introduction
Tenacibaculum maritimum (formerly Flexibacter maritimus), a member of the family Flavobacteriaceae, phylum Bacteroidetes (Suzuki et al., 2001), is the etiological agent of tenacibaculosis, a very serious bacterial disease of many commercial marine fish species (for a review, see Avendaño-Herrera et al., 2006b), responsible for considerable economic losses in all major areas of marine finfish aquaculture worldwide (i.e., Japan, Europe including the Atlantic, Channel and Mediterranean coasts, North America, Australia, and the Red Sea). Moreover, T. maritimum can affect a large number of feral, captive, and cultured fish species such as: Dover sole (Solea solea), Senegalese sole (Solea senegalensis), wedge sole (Dicologoglossa cuneata), turbot (Scophthalmus maximus), Atlantic salmon (Salmo salar), Japanese flounder (Paralichthys olivaceus), yellowtail (Seriola quinqueradiata), red sea bream (Pagrus major), black sea bream (Acanthopagrus schlegelii), gilthead sea bream (Sparus aurata), European sea bass (Dicentrarchus labrax), puffer fish (Takifugu rubripes), Pacific sardine (Sardinops sagax), lumpsucker (Cyclopterus lumpus), and sand tiger shark (Carcharias taurus) (Bernardet et al., 1990; Avendaño-Herrera et al., 2006a; López et al., 2009; AbdEl-Galil and Hashiem, 2011; Rahman et al., 2014; Florio et al., 2016; Småge et al., 2016; and references therein]. Affected fish usually display a variety of external signs including eroded mouth, skin ulcers, fin necrosis, and tail-rot. Skin lesions are often colonized by opportunistic pathogens such as Vibrio spp. So far, only one specific vaccine is commercially available to prevent tenacibaculosis in turbot. Hence, in all other fish species, the control of tenacibaculosis outbreaks remains restricted to the use of antibiotics, sometimes combined with external disinfectants (Avendaño-Herrera et al., 2008).
So far, three serotypes have been documented that show varying degrees of association with host fish species (Avendaño-Herrera et al., 2005a). This serological diversity could have important consequences for the development of an efficient vaccine. Recently, multilocus sequence analysis (MLSA) of T. maritimum isolates representative of the worldwide diversity revealed that this species constitutes a cohesive group, exhibiting moderate levels of nucleotide diversity and recombination [average pairwise nucleotide diversity (π) estimated to be 0.44% and r/m ratio estimated to be 2.7]. Moreover, the population structure of T. maritimum did not reveal dominant genotypes or clonal complexes but rather suggested an endemic colonization of fish farms by local strains with no contribution of long-distance contamination related to fish movements. In addition, the same MLSA genotype was identified in different host species in the same geographical area, suggesting host versatility (Habib et al., 2014).
Despite the significance of tenacibaculosis outbreaks in the aquaculture industry, little is known about the virulence mechanisms of T. maritimum (Avendaño-Herrera et al., 2006b). Adhesion to hydrophobic surfaces (Burchard et al., 1990) or fish skin mucus (Magariños et al., 1995), hemagglutination (Pazos, 1997), extracellular products including proteolytic activity (Baxa et al., 1988; Handlinger et al., 1997; Pazos, 1997; van Gelderen et al., 2009), and iron uptake mechanisms (Avendaño-Herrera et al., 2005b) have been suggested to play roles in virulence. However, the molecular factors involved remain to be identified. Loss-of-function studies for experimental validation of genes as virulence factors are still inaccessible due to the absence of genetic tools.
In the present work, we sequenced and analyzed the complete genome of T. maritimum NCIMB 2154T to forecast the genes relevant to the bacterial lifestyle, in particular those linked to virulence. These in silico predictions paved the way for assessing for the first time the functional role of some relevant components. This genome will serve as a reference for future whole genome-based molecular epidemiology surveys aimed at analyzing disease emergence and propagation (Bayliss et al., 2017).
Materials and Methods
Bacterial Growth Conditions
Several batches of the T. maritimum type strain (i.e., NCIMB 2154T, ATCC 43398T, CIP 103528T, and DSM 17995T), Tenacibaculum discolor LL04 11.1.1T, Tenacibaculum jejuense CNURIC013T, and Tenacibaculum soleae LL04 12.1.7T were routinely grown in marine broth and agar 2216 (Difco) at 28°C and 170 rpm.
Genome Sequencing
Tenacibaculum maritimum NCIMB 2154T was sequenced with a combination of PacBio RSII (N50 reads 7.4 kb, estimated coverage 234 x) and Illumina (HiSeq 2x100 pair-end reads with 300 bp insert size, 54,259,876 filtered sequences, estimated coverage 1500 x) reads and assembled with MHAP to completion to obtain a circular molecule. The final, quiver polished assembly was validated by optical mapping using NcoI.
Annotation and Genome Comparisons
Genome annotation, including manual curation, and comparisons were performed using the web interface MicroScope (Vallenet et al., 2013) which allows graphic visualization enhanced by a synchronized representation of synteny groups1. Predictions of repeated sequences were performed using Repseek (Achaz et al., 2007) and those of genomic islands (GIs) using SIGI-HMM (Waack et al., 2006) and Alien hunter (Vernikos and Parkhill, 2006). The dbCAN database was used to identify carbohydrate active enzymes (CAZymes)2 (Yin et al., 2012). The genomic sequence reported in this article has been deposited in the EMBL database under the accession number LT634361.
MLST on Selected Strains
The four above-mentioned batches of the T. maritimum type strain were genotyped using the MLST scheme described in Habib et al. (2014).
Chondroitin AC Lyase and Sphingomyelinase Cloning, Expression, and Enzymatic Activity
The genes encoding the chondroitin AC lyase (cslA, locus identifier: MARIT_2107) and sphingomyelinase (sph, locus identifier: MARIT_1748) were cloned according to Groisillier et al. (2010). Briefly, primers were designed to amplify the coding region corresponding to the catalytic module of CslA (forward primer 5′-TTTTTTAGATCTACTTCTCTAACTTTGGATGTAAATTCG-3′; reverse primer 5′-TTTTTTGAATTCTTATATTTTAAGAACTTTCTCTGTTATTAG-3′) and sph (forward primer 5′-AAAAAAGGATCCAATGATGACGTTTCCCTTGGAGAAA-3′; reverse primer 5′-TTTTTTCAATTGTTAGTAGCTAAAGTAAAAAGTTTGCTTG-3′) by PCR from T. maritimum genomic DNA. After digestion with the restriction enzymes BglII and EcoRI, and BamHI and MfeI respectively, the purified PCR products were ligated using the T4 DNA ligase into the expression vector pFO4 predigested by BamHI and EcoRI (referred to as the plasmid pCslA and psph), resulting in a recombinant protein with a N-terminal hexa-histidine tag for each construct. The obtained plasmids were transformed into Escherichia coli DH5α for storage and in E. coli BL21(DE3) for protein expression. E. coli BL21(DE3) cells harboring the plasmid pCslA or psph were cultivated at 20°C in a 3 mL auto-induction ZYP 5052 medium (Studier, 2005) supplemented with 100 μg/mL ampicillin. Cultures were stopped after 72 h and centrifuged for 35 min at 4°C, 3,000 g. The cells were resuspended in 500 μL of buffer A (20 mM sodium phosphate pH 7.4, 500 mM NaCl, 10 mM imidazole). An anti-proteases mixture (cOmpleteTM EDTA-free, Roche) and 0.1 mg/mL of DNase were added. The cells were disrupted by sonication. After centrifugation at 12,500 g for 2 h at 4°C the supernatant was loaded onto a His spin trap column (GE Healthcare Life Science) equilibrated with buffer A. After extensive washing with buffer A, the recombinant proteins were eluted with 400 μL of buffer B (20 mM sodium phosphate pH 7.4, 500 mM NaCl, 500 mM imidazole). The results were analyzed by 12 % sodium dodecyl sulfate-polyacrylamide gel electrophoresis.
Chondroitin AC Lyase In Vitro Activity
The native chondroitin AC lyase activity of T. maritimum was assayed in vitro according to Li et al. (2015). Briefly, 6 μL of mid-log-phase bacterial cultures (OD600 = 0.6) were spotted on marine agar 2216 supplemented with 0.2% chondroitin sulfate A or C (Sigma) and 2% bovine serum albumin (BSA, Sigma), and incubated for 48 h at 28°C. Chondroitin lyase activity was visualized as a clear halo surrounding the bacterial growth after the plates were flooded with 0.35 N HCl.
The activity of the recombinant CslA was determined by adding 1 μL of the recombinant protein solution to a 1-mL cuvette containing 600 μL of 50 mM Tris–HCl, pH 8.0 supplemented by 1 mg/mL of chondroitin A or C at 30°C. Product formation was monitored as an increase in absorbance at 232 nm as a function of time (Michel et al., 2004). The assay was performed in triplicate.
Sphingomyelinase Activity
The activity of the recombinant Sph was determined using a coupled assay Amplex Red Sphingomyelinase assay kit (Life Technologies, Invitrogen) following the manufacturer’s instructions. The reactions were performed in 96-well special optics flat clear bottom black polystyrene Microplates (Corning). The reaction mixture (200 μL) contained 100 μL of 1.3 and 13 ng of recombinant protein and 100 μL of 100 μM Amplex red reagent (containing 2 U/mL horseradish peroxidase, 0.2 U/mL choline oxidase, 8 U/mL alkaline phosphatase, and 0.5 mM sphingomyelin). The fluorescence was measured every minute at excitation and emission wavelengths of 530 nm and 590 nm, respectively, using the TECAN Infinite® Pro200 microplate reader at 28°C for 15 min. The background fluorescence was corrected by subtracting the negative control (i.e., without recombinant protein). The positive control of each experiment was performed with Bacillus cereus sphingomyelinase provided by the manufacturer. The assay was performed in triplicate.
Sialidase Activity
The fluorogenic substrate 2′-(4-methylumbelliferyl)-α-D-N-acetylneuraminic acid sodium salt hydrate (MUAN) was used to determine sialidase activity according to Mally et al. (2008). Briefly, 107 mid-log phase bacteria were incubated with 0.1 mM of MUAN (Sigma) in 100 mM sodium acetate buffer pH 7.4 at 28°C. The reaction was stopped by the addition of a 0.5 M Na2CO3 solution at pH 10. Released 4-methylumbelliferone was measured by fluorescence in a TECAN Infinite® Pro200 microplate reader at an excitation wavelength of 360 nm and an emission wavelength of 440 nm. The background fluorescence was corrected by subtracting the negative control (i.e., without bacteria). The assay was performed in triplicate.
Results and Discussion
General Genome Features
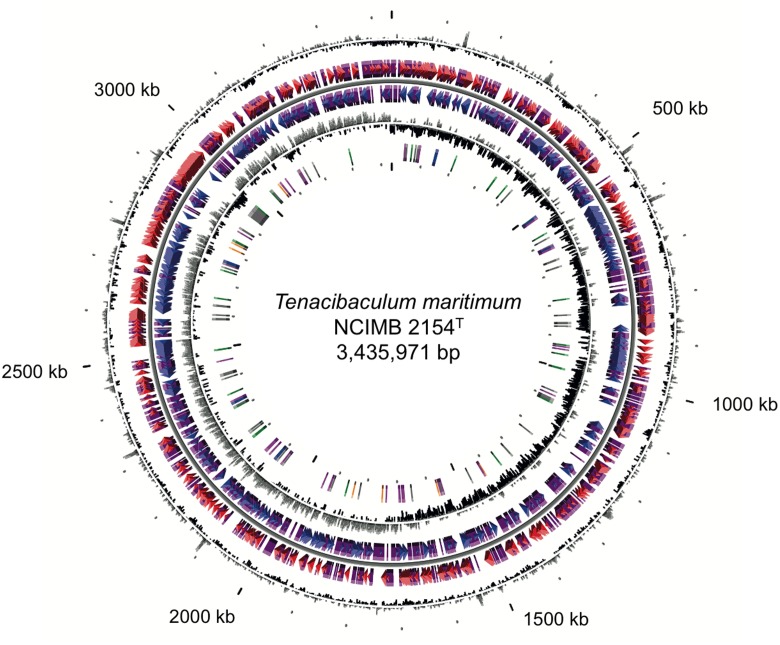
The genome of T. maritimum NCIMB 2154T consists of a circular chromosome of 3,435,971 bp with a 32.01% GC content. No plasmid was identified. The chromosome is predicted to contain 2,866 protein-coding genes, 6 rDNA operons, and 57 tRNA (Figure 1). The minimal gene set includes well-conserved housekeeping genes for basic metabolism and macromolecular synthesis, many of which are essential (the list of these genes was taken from Gil et al., 2004). Accordingly, we formally identified 203 out of the 206 protein-coding genes proposed by these authors to represent this core minimal gene set. Strikingly, the genome is rich in repeat sequences, encompassing 10.35% of the genome. Twenty-five GIs were predicted (Supplementary Table S1), two of which (GI2 and GI9) are likely from phage origin (i.e., containing elements such as a tRNA border and motility genes, and displaying a compositional bias). Genes encoding rhs family proteins and rhs (rearrangement hot-spot) associated vgr proteins (or their remnants) were frequently identified in the predicted islands. Genes encoding Cas1, Cas2, and Cas9 were identified in GI19. However, the unconventional gene organization of the cas genes and the lack of short direct repeats interspersed with spacer sequences strongly argues for a functionally defective CRISPR system.
FIGURE 1.
Circular representation of the Tenacibaculum maritimum NCIMB 2154T genome. Circles display (from the outside): (1) GC percent deviation (GC window – mean GC) in a 1,000-bp window. (2) Predicted CDSs transcribed in the clockwise direction. (3) Predicted CDSs transcribed in the counterclockwise direction. Genes displayed in (2) and (3) are color-coded according to the following categories: red and blue, MaGe validated annotations; purple, primary/automatic annotations. (4) GC skew (G + C/G – C) in a 1,000-bp window. (5) rRNA (blue), tRNA (green), miscellaneous other RNA (orange), transposable elements (pink), and pseudogenes (gray).
The Genome of T. maritimum NCIMB 2154T Reveals Errors in Culture Collections
While this work was in progress, a WGS assembly of the T. maritimum type strain retrieved from the NBRC collection (NBRC 15946T; RefSeq assembly accession: GCF_000509405.1) was released but not published. Its comparison with the complete genome of strain NCIMB 2154T presented in this study revealed unexpected sequence discrepancies as both cultures were supposed to represent the type strain. Using MLST (Habib et al., 2014), we confirmed that strains NCIMB 2154T, ATCC 43398T, and CIP 103528T all share identical sequences for the seven loci [corresponding to Sequence Type 1 (ST1)] and are, as predicted, most likely of the same origin. In contrast, strain NBRC 15946T and its derivative strain DSM 17995T, both possess the same sequences for the seven loci (corresponding to ST32) that differ from the sequences of ST1 at four loci (i.e., atpA, dnaK, glyA, and gyrB). According to the dates and order of deposition in the different culture collections, NCIMB 2154T, ATCC 43398T, and CIP 103528T represent the bona fide type strain (Supplementary Figure S1).
Metabolism
Genome analysis indicated the presence of a complete Embden–Meyerhof–Parnas pathway, of the tricarboxylic acid cycle and of genes encoding NADH dehydrogenase subunits, cytochrome c, cytochrome c oxidase, components of ATP synthase genes as well as enzymes needed to synthesize amino acids, nucleotides, fatty acids, heme, vitamins, and coenzymes (e.g., biotin, farnesyl diphosphate, coenzyme A, NAD, FAD, dihydrofolate, mevalonate, and thiamin). However, and in contrast with T. soleae, the cobalamin biosynthesis encoding genes are absent in the T. maritimum genome. Nitrate reduction (Wakabayashi et al., 1986) is likely performed by the periplasmic, cytochrome c-linked, nitrate reductase complex NapAB (MARIT_1701-1700).
A relevant characteristic of the lifestyle of pathogens is nutrient acquisition from their host. T. maritimum is able to degrade proteinaceous compounds (e.g., gelatin and casein) and to grow on casamino acids or tryptone as a sole carbon and nitrogen source (Wakabayashi et al., 1986). Accordingly, the T. maritimum genome encodes predicted secreted proteases (see below), peptide/amino acid transporters and peptide/amino acid catabolic pathways likely involved in protein degradation and uptake from host tissues. T. maritimum has been reported to be unable to degrade most simple and more complex carbohydrates (Wakabayashi et al., 1986; Avendaño-Herrera et al., 2006b). However, sugar transporters and CAZymes were predicted. Overall, the genome of T. maritimum encodes 59 CAZymes encompassing 18 glycosyl hydrolase, 30 glycosyl transferases, one polysaccharide lyase, six carbohydrate binding modules containing proteins, and four carbohydrate esterases (Supplementary Table S2).
Polysaccharide utilization loci (PUL), restricted to and very common within the phylum Bacteroidetes, are gene clusters involved in the capture, degradation, and import of complex carbohydrates. PUL-encoded proteins encompass a SusD-family cell-surface lipoprotein that binds the oligosaccharide, and a SusC-family TonB-dependent receptor for its transport across the bacterial outer membrane (Anderson and Salyers, 1989). Moreover, genes encoding SusC- and SusD-family proteins are usually organized in tandem in the genomes of Bacteroidetes (Terrapon et al., 2015). In full accordance with its inability to use carbohydrates, the T. maritimum genome presents a very low amount of susC/susD pairs compared to other members of the phylum Bacteroidetes (Barbeyron et al., 2016). Moreover, among the six identified susC/susD containing loci, only one harbors a typical PUL structure (MARIT_2678 - 2679) and is predicted to be involved in glycan harvesting from host glycoproteins (see below).
Iron Acquisition and Utilization
Iron acquisition from host plays an important role in virulence of many pathogenic bacteria. In biological systems, high-affinity iron-binding proteins can chelate iron, and pathogens have developed efficient mechanisms to obtain iron from their hosts (Ratledge and Dover, 2000). In the T. maritimum genome, we identified a siderophore biosynthesis gene cluster (MARIT_0169-0174) highly similar to the mbs locus from a deep-sea metagenome (Fujita et al., 2012). As this gene cluster is predicted to be involved in the production of the macrocyclic hydroxamate class bisucaberin siderophore, we named the genes tbs for Tenacibaculum bisucaberin synthase. However, the gene organization is different from those previously reported displaying a major facilitator-family exporter-encoding gene and a duplication/fusion of the tbsCD gene (Supplementary Figure S2). A highly similar locus is observed in the genome of the Tenacibaculum mesophilum type strain (CIP 107215T; data not shown), likely responsible of the bisucaberin B siderophore biosynthesis as proposed by Fujita et al. (2013). Among the numerous TonB-dependent outer membrane receptors encoded in the T. maritimum genome, MARIT_0185, located in the tbs locus neighborhood, likely encodes the bisucaberin siderophore-iron transporter.
In the human periodontal bacterium Porphyromonas gingivalis, the heme-binding lipoprotein HmuY, together with the outer-membrane receptor HmuR, are predicted to be virulence factors during bacterial infection (Olczak et al., 2008; Wójtowicz et al., 2009). In the T. maritimum genome, two genes (MARIT_1312-1313), organized in tandem, encode HmuR and HmuY homologous proteins and are predicted to be involved in heme uptake.
In addition, MARIT_0141 – 0142 encoding FeoAB likely constitute a Fe2+ uptake system (Lau et al., 2016) and two iron-regulated protein homologous genes (MARIT_1664 and MARIT_1661), belonging to the imelysin family, might also be involved in iron acquisition, uptake or storage (Xu et al., 2011). The control of iron metabolism is likely carried out by the ferric uptake regulator Fur (MARIT_1835; Fillat, 2014). Hence, such a variety of iron acquisition systems strongly suggests the ability of this bacterium to survive under poor iron conditions (sea water) and/or to retrieve iron sequestered by host proteins (Avendaño-Herrera et al., 2005b).
Motility, Adhesion, Quorum Sensing/Quenching, and Stress Response
Like most members of the family Flavobacteriaceae, T. maritimum moves over surfaces by gliding motility, an active process that does not involve pili or flagella. The genome of T. maritimum encodes all the proteins that form the gliding machinery, i.e., the 14 gld genes (gldA to gldN; McBride et al., 2009) and 10 spr genes (sprA, sprB, sprC, sprD, sprE, and five sprF paralogs; McBride and Zhu, 2013). T. maritimum NCIMB 2154T is extremely adherent to different surfaces including agar, plastic, and glass. Genes encoding (i) the biosynthesis of exopolysaccharides (MARIT_2522-2537); (ii) the numerous adhesins (n = 17); and (iii) the proteins displaying lectin or carbohydrate-binding motifs could be involved in these strong adhesive properties, in the biofilm-forming ability and in the hemagglutination properties of the bacterium (Pazos, 1997).
Quorum sensing is a bacterial communication process that controls a range of functions at the population level. In Gram-negative bacteria, the most studied quorum sensing system comprises the production and detection of acyl homoserine lactones (AHLs), diffusible compounds that act as signaling molecules between cells (Garg et al., 2014). Though AHL production was previously reported in T. maritimum (Romero et al., 2010), no homologous gene for AHL biosynthesis was detected in its genome. In contrast, quorum quenching refers to all processes involved in the inhibition of bacterial communication (Kalia, 2013). A N-acyl homoserine lactonase encoding gene (GenBank: KR232938.1) belonging to the metallo-β-lactamase family has been proposed to be the quorum quencher of T. maritimum (Mayer et al., 2015). However, this gene is definitively absent from the T. maritimum genome and one must conclude that KR232938.1 does not belong to T. maritimum but rather to another fish pathogen, T. discolor (99.77 % nucleotide sequence identity).
Pathogenic bacteria have to adapt to the changing environments between their different lifestyles and to cope with various stresses including reactive oxygen species (ROS) produced by host macrophages. The genome of T. maritimum encodes three superoxide dismutases (SodA, SodB, and SodC). Most bacteria possess either a manganese-dependent (SodA) or an iron-dependent (SodB) superoxide dismutase in their cytoplasm, while zinc-dependent superoxide dismutases (SodC) have been detected mostly in pathogenic bacteria (Sheng et al., 2014). These enzymes convert superoxide anions to molecular oxygen and hydrogen peroxide, to be further metabolized by catalases or peroxidases. The presence of the three types of superoxide dismutases and two catalase/peroxidase (KatA and KatG) suggests that T. maritimum uses a sophisticated mechanism to face up oxidative stress. In addition, three loci involved in bacterial resistance to heavy metals have been identified: (i) MARIT_0364-0366, similar to the drug efflux system AcrA–AcrB–TolC of E. coli (Lee et al., 2012); (ii) MARIT_1200 encoding a putative arsenate reductase; and (iii) MARIT_1768-1771 encoding a heavy metal efflux pump-type ATPase. These loci are likely involved in the removal of cationic heavy metals to limit the production of ROS by the Fenton reaction.
Transport and Secretion Systems
Transport systems are of great significance for virulence by addressing toxins to the bacterial surface. ABC-type transport systems, the Sec-dependent transport system, and the twin-arginine transport system were identified. In the phylum Bacteroidetes, the type IX secretion system (T9SS) allows the delivery of proteins to the cell surface (McBride and Zhu, 2013). All previously characterized components of the T9SS were identified in the T. maritimum genome. In P. gingivalis, the T9SS-secreted proteins comprise many virulence factors, including the extracellular and cell-surface cysteine proteinases gingipains (Sato et al., 2010, 2013). These T9SS-secreted proteins possess a conserved C-terminal domain (CTD), involved in secretion and cell-surface anchoring. These 70–100 amino acids long CTDs belong to two different TIGRFAM protein domain families, TIGR04183 and TIGR04131 (McBride and Nakane, 2015). The T. maritimum genome encompasses eight genes encoding TIGR04131-containing proteins (Supplementary Table S3). Most, if not all, are predicted to be adhesins, including SprB (MARIT_1321), which is also required for gliding motility (Nelson et al., 2008). In addition, several predicted toxins were identified among the 43 genes encoding TIGR04183-containing proteins.
Toxins
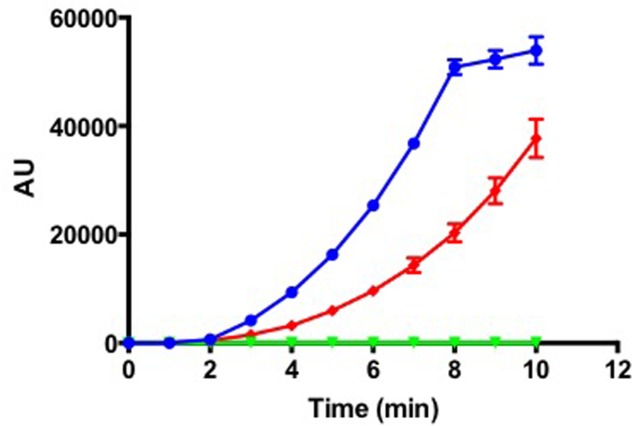
As T. maritimum is a pathogenic bacterium, this species should possess sophisticated mechanisms to invade and colonize host tissues. Accordingly, the T. maritimum genome encodes a bunch of predicted toxins and virulence factors including membrane-damaging enzymes potentially involved in host cells lysis. A gene encoding a sphingomyelinase with a lipoprotein signal (MARIT_1748) homologous (30.2% identity/50% similarity) to the one of B. cereus was identified (Supplementary Figure S3A). A gene encoding a ceramidase with a signal peptide and a TIGR04183 domain (MARIT_2033) homologous (33.5% identity/59.8% similarity) to the one of Pseudomonas aeruginosa (Supplementary Figure S3B) is also present in the T. maritimum genome. Sphingomyelinase has been reported to be cytotoxic to host cells by acting as a potent hemolytic factor (Oda et al., 2010), while the bacterial ceramidase functions as an exotoxin or activator of exotoxin (Okino et al., 2010; Ito et al., 2014). Indeed, the outer layer of the plasma membrane of eukaryotic cells contains phospholipids, which are hydrolyzed to phosphocholine and ceramide by sphingomyelinase, the latter being subsequently hydrolyzed to sphingosine and fatty acids by a ceramidase. To formally demonstrate that MARIT_1748 encodes the sphingomyelinase, we cloned the corresponding nucleotide sequence in the pFO4 vector (Groisillier et al., 2010). The recombinant protein was produced in a soluble form in E. coli BL21(DE3) and the enzymatic activity of the purified sphingomyelinase was assayed in triplicate using the Amplex Red Sphingomyelinase assay kit (Figure 2). Another predicted hemolysin is encoded by MARIT_0124 and belongs to the cholesterol-dependent cytolysin family (Gilbert, 2010). These pore-forming toxins were originally identified in Gram-positive bacteria and encompass well-known examples including listeriolysin, perfringolysin, streptolysin, and pneumolysin (Los et al., 2013).
FIGURE 2.
Tenacibaculum maritimum sphingomyelinase activity. Fluorescence measurement (arbitrary units) following incubation of 1.3 ng (blue line) of recombinant protein with Amplex Red Sphingomyelinase assay kit. The control conditions were as follows: negative control, 1.3 ng of boiled recombinant protein (green line); positive control, the purified sphingomyelinase from Bacillus cereus provided by the manufacturer (red line). Results correspond to the mean of triplicates and SDs are included.
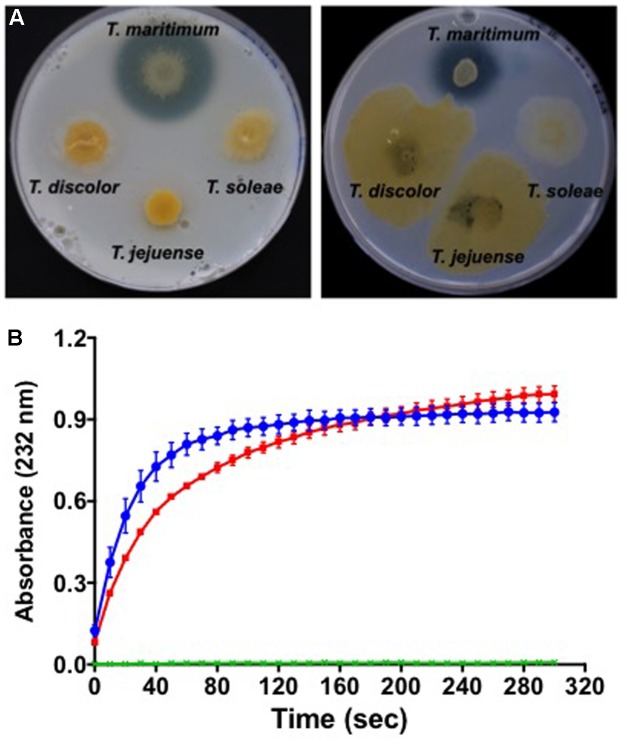
Glycosaminoglycans (GAGs) are highly sulfated polymers composed of repeated disaccharide units (an amino sugar and an uronic sugar). They represent major components of animal cell surface and extracellular matrix, mostly in the form of proteoglycans. Among them, chondroitin sulfate is an important component of cartilage and fish connective tissue (Arima et al., 2013). It is composed of a chain of alternating N-acetylgalactosamine and glucuronic acid to which proteins attach. Chondroitin sulfate lyases have been suggested to be virulence factors, for instance in the other fish pathogen Flavobacterium columnare (Suomalainen et al., 2006). One might predict that the cslA gene (MARIT_2107) encoding a PL8_3 family chondroitin AC lyase highly similar to that of F. columnare plays a similar role. In vitro analyses demonstrated that T. maritimum is able to degrade chondroitin sulfate A and C on marine agar 2216, as showed by the formation of a degradation halo around the bacterial growth (Figure 3A). Although a chondroitin C-lyase activity was recently suggested for T. maritimum (Rahman et al., 2014), our results demonstrate the ability of this bacterium to also degrade chondroitin sulfate A. Other phylogenetically close Tenacibaculum species, such as T. discolor, T. jejuense, or T. soleae do not display this chondroitin AC lyase activity under the same conditions. To formally demonstrate that gene cslA encodes the chondroitin AC lyase, we cloned the nucleotide sequence corresponding to the PL8_3 catalytic module in the pFO4 vector (Groisillier et al., 2010). The recombinant protein, referred to as TmCslAPL8, was produced in a soluble form in E. coli BL21(DE3). The enzymatic activity of the purified TmCslAPL8 was assayed in triplicate by measuring the increase in absorbance at 232 nm of the reaction products using chondroitin A and C sulfates as substrates. As seen in Figure 3B, TmCslAPL8 is highly active on both substrates, confirming the functional annotation of MARIT_2107. As no sulfatase could be identified in the genome, it is likely that GAGs such as chondroitin sulfate cannot be assimilated by T. maritimum. Therefore, CslA might be a bona fide virulence factor allowing the pathogen to invade fish tissues.
FIGURE 3.
Degradation of chondroitin sulfates by T. maritimum. (A) Colonies of T. maritimum NCIMB 2154T, T. discolor LL04 11.1.1T, T. jejuense KCTC 22618T, and T. soleae LL04 12.1.7T on marine agar 2216 supplemented with 0.2% of chondroitin sulfates A (left) and C (right). (B) Activity of the recombinant protein TmCslAPL8 on chondroitin A (blue line) and chondroitin C (red line). The release of unsaturated oligosaccharides was monitored by spectrophotometry at 232 nm using biological triplicates. The two control conditions (green lines) correspond to the same reaction mixture with a boiled TmCslAPL8. For the clarity of the graphic, the error bars are only indicated every 10 s.
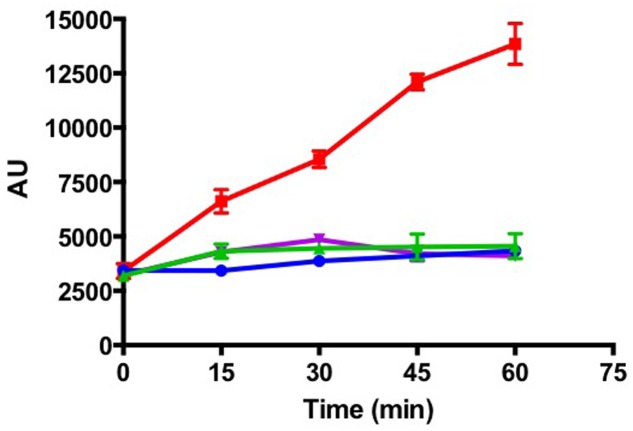
Capnocytophaga canimorsus, another member of the family Flavobacteriaceae, is a commensal of cat and dog mouth that can cause dramatic infections in bitten humans (Pers et al., 1996). C. canimorsus has the unusual property to feed directly on cultured mammalian cells by harvesting the glycan moiety of cellular glycoproteins, a property dependent on SiaC (Mally et al., 2008). Sialic acids are predominantly found in cell-surface exposed and secreted eukaryotic glycoproteins, being involved in many physiological, biological, and immunological functions (Varki and Varki, 2007). Mucosal surfaces are especially sialoglycan-rich and bacterial sialidases play important roles during the colonization and damage of mammalian mucosal surfaces (Lewis and Lewis, 2012). In T. maritimum, siaA (MARIT_2686) encodes a predicted sialidase, which activity was formally demonstrated using the fluorogenic substrate MUAN (Figure 4). In contrast, other Tenacibaculum species including the two fish pathogens T. soleae and T. discolor, for which no siaA homologous gene has been identified (data not shown), were unable to degrade MUAN under the same experimental conditions. Indeed, the siaA gene is encompassed in a [GI N° 24 inserted in a Gln-tRNA (position 2,965,888), Supplementary Table S1] and has a predicted foreign origin. The 3′ part of this GI (2,916,321–2,965,888) is mainly composed of pseudogenes including scars of transposases, Vgr family proteins and Rhs family proteins. On the other hand, the 5′ part of this GI (2,916,321–2,915,448) contains nine bona fide genes, predicted to encode a PUL encompassing (i) a SusC/SusD outer-membrane importer system; (ii) a N-acetylneuraminate lyase; (iii) a N-acyl-D-glucosamine 2-epimerase; and (iv) a N-acetylneuraminate epimerase/sodium:sialic acid symporter-fusion inner-membrane protein. This PUL system is likely dedicated to the harvesting, import, and catabolism of sialic acids from host glycoproteins (Supplementary Figure S4). Intriguingly, the sialidase displays a very unique structure including (i) a signal peptide; (ii) a carbohydrate esterase family 6 domain; (iii) a family 40 carbohydrate-binding module; (iv) two adjacent, fully duplicated, family 33 glycoside hydrolase domains; and (v) a TIGR04183 domain for T9SS-mediated secretion and cell-surface anchoring. Moreover, the predicted mechanism of glycan harvesting by T. maritimum is likely different from the one identified in C. canimorsus. Indeed, the T. maritimum genome is devoid of the gpdCDGEF operon encoded by the C. canimorsus PUL5 and involved in deglycosylation and import of N-linked oligosaccharides (Renzi et al., 2011). In addition, the C. canimorsus sialidase is a periplasmic-exposed lipoprotein that processes oligosaccharides after SucC/SusD-mediated import, whereas the T. maritimum sialidase is predicted to be cell-surface exposed, therefore directly processing sialic acid from glycoproteins. Hence, one might predict that both species perform the same function (foraging host glycoproteins) using common strategies (sialidase, PUL) but in sequentially different ways.
FIGURE 4.
Tenacibaculum maritimum sialidase activity. Fluorescence measurement (arbitrary units) following incubation of T. maritimum NCIMB 2154T (red line), T. discolor LL04 11.1.1T (blue line), T. jejuense KCTC 22618T (green line), and T. soleae LL04 12.1.7T (purple line) cells with the fluorogenic substrate MUAN. Results correspond to the mean of triplicates and SDs are shown.
Extracellular proteases may exhibit a wide range of virulence potentials when interacting with the host defense mechanisms and tissue components. Furthermore, they may promote the survival of pathogens under adverse environmental conditions encountered in the infected host (Dubin, 2002). Since early studies, T. maritimum was shown to be proteolytic (Wakabayashi et al., 1986) and its proteases were suspected to act synergistically with other virulence factors, leading to tissue destruction and mortality (Baxa et al., 1988). Eight cell-surface-exposed, TIGR04183 domain-containing proteases were predicted (Supplementary Table S3), two of which likely of importance. MARIT_2328 encodes a multi-domain protein encompassing a C10 family peptidase, highly similar to streptopain (SpeB), an important streptococcal virulence factor likely playing a role in bacterial colonization, invasion, and inhibition of wound healing (Nelson et al., 2011). MARIT_1085 encodes a collagenase similar to that of Cytophaga sp. strain L43-1 (Sasagawa et al., 1995).
Conclusions and Perspectives
We report here the complete genome sequence of T. maritimum, a serious pathogen of marine fish in many geographical areas. T. maritimum shows a lack of host specificity, affecting a variety of wild and farmed fish species (Avendaño-Herrera et al., 2006b). Sequence analysis has revealed a combination of strategies that probably confers T. maritimum the ability to invade, colonize, and degrade fish tissues and to exploit some cellular compounds for growth. The central metabolism of T. maritimum is similar to that of the other flavobacteria sequenced to date (e.g., several Flavobacterium, Gramella, Dokdonia, and Polaribacter species). However, T. maritimum does not possess a proteorhodopsin-encoding gene as identified in close relatives such as Polaribacter, Dokdonia, or Psychroflexus species, suggesting the inability of this bacterium to use light to generate proton motive force. Comparison with the available genomes of the three other fish-pathogenic Tenacibaculum species Tenacibaculum dicentrarchi, Tenacibaculum ovolyticum, and T. soleae (Grothusen et al., 2016; Lujan et al., 2016; Teramoto et al., 2016), has revealed striking differences in virulence strategies as most, if not all, the aforementioned predicted toxins (Table 1) are absent from the genomes of the latter species. These elements point to very different paths in the evolution of virulence as suggested using a subset of core-genome genes (Habib et al., 2014). The genome sequence of T. maritimum provides insights into the lifestyle of this poorly studied pathogen and may help in the development of efficient control strategies in fish farms. Indeed, the predicted virulence factors could lead to the development of attenuated T. maritimum variants for vaccine development. The genome of the type strain may also serve as a reference for future genomic comparisons for a better understanding of intraspecies and intragenus diversity and evolution as well as whole genome-based molecular epidemiology studies (Bayliss et al., 2017).
Table 1.
Summary of the predicted virulence-associated genes identified in this study in the genome of T. maritimum NCIMB 2154T.
| Label | Predicted function | Activity present in the following species | Activity suspected in T. maritimum |
|---|---|---|---|
| Iron uptake | |||
| MARIT_0169-0174 | Siderophore biosynthesis system, Tbs | Deep sea metagenome (Fujita et al., 2012) | Avendaño-Herrera et al., 2005b |
| MARIT_1312-1313 | Heme uptake mechanism, HmuYR | Porphyromonas gingivalis (Olczak et al., 2008; Wójtowicz et al., 2009) | Avendaño-Herrera et al., 2005b |
| MARIT_0141 – 0142 | Fe2+ uptake system, FeoAB | Porphyromonas gingivalis (Lau et al., 2016) | Avendaño-Herrera et al., 2005b |
| MARIT_1664 | Iron acquisition, uptake or storage, imelysin family protein | Synechococcus elongatus (Xu et al., 2011) | Avendaño-Herrera et al., 2005b |
| MARIT_1661 | Iron acquisition, uptake or storage, imelysin family protein | Synechococcus elongatus (Xu et al., 2011) | Avendaño-Herrera et al., 2005b |
| Stress resistance | |||
| MARIT_3105 | Superoxide dismutase [Mn/Fe], SodA | Staphylococcus carnosus (Barrière et al., 2001) | Suzuki et al., 2001 |
| MARIT_1670 | Superoxide dismutase [Fe], SodB | Legionella pneumophila (Amemura-Maekawa et al., 1996) | Suzuki et al., 2001 |
| MARIT_1821 | Superoxide dismutase [Cu–Zn], SodC | Schistosoma mansoni (da Silva et al., 1992) | Suzuki et al., 2001 |
| MARIT_0946 | KatG catalase | Geobacillus stearothermophilus (Singh et al., 2008) | Suzuki et al., 2001 |
| MARIT_2408 | KatA catalase | Pseudomonas aeruginosa (Brown et al., 1995) | Suzuki et al., 2001 |
| Toxins | |||
| MARIT_0124 | Cholesterol-dependent cytolysin | Capnocytophaga canimorsus (Gilbert, 2010) | Baxa et al., 1988 |
| MARIT_1085 | Collagenase | Cytophaga sp. (Sasagawa et al., 1995) | Baxa et al., 1988 |
| MARIT_1748 | Sphingomyelinase | Bacillus cereus (Oda et al., 2010) | |
| MARIT_2033 | Ceramidase | Pseudomonas aeruginosa (Ito et al., 2014) | |
| MARIT_2107 | Chondroitin AC lyase | Flavobacterium columnare (Suomalainen et al., 2006) | Rahman et al., 2014 |
| MARIT_2328 | Streptopain family protease | Streptococcus pyogenes (Nelson et al., 2011) | Wakabayashi et al., 1986; Baxa et al., 1988 |
| MARIT_2678 - 2687 | Sialoglycan degradation and uptake | Capnocytophaga canimorsus (Mally et al., 2008) | |
Author Contributions
DP-P performed genome annotation, phenotypic characterization, and drafting the manuscript; AL, AR, and CL-R performed DNA extraction, library construction, and sequencing; GhM performed the optical-mapping; ZR participated in genomic data analysis; RL, TB, AG, and GuM performed gene cloning, protein expression, and biochemical characterization with substantial intellectual contribution; J-FB substantial intellectual contribution throughout the study, data analysis, and manuscript preparation. ED substantial intellectual contribution throughout the study, gene mining, interpretation of data, manuscript preparation, and responsible for acquisition of funding. All authors read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are thankful to Institut Français de Bioinformatique (IFB), V. Barbe (CEA/Genoscope) for the optical mapping, V. Loux (MaIage) from the INRA MIGALE bioinformatics platform (http://migale.jouy.inra.fr) for help with bioinformatics and C. Klopp (GenoToul Bioinfo) for his contribution to genome assembly.
Funding. This work was supported by the Agence Nationale pour la Recherche (contract ANR-14-CE19-0020). It has benefited from the expertise of the High-throughput Sequencing platform of I2BC (http://www.i2bc.paris-saclay.fr) and GeT core facility platform (http://get.genotoul.fr) and was supported by the France Génomique national infrastructure, funded as part of “Investissement d’avenir” program managed by Agence Nationale pour la Recherche (contract ANR-10-INBS-09).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.01542/full#supplementary-material
References
- AbdEl-Galil M. A. A., Hashiem M. (2011). Tenacibaculosis in Picasso tigger fish (Rhinecanthus assasi) and black damsel fish (Neoglyphieodon meles) of Red sea at Hurghada, Egypt. Life Sci. J. 8 1166–1171. [Google Scholar]
- Achaz G., Boyer F., Rocha E. P., Viari A., Coissac E. (2007). Repseek, a tool to retrieve approximate repeats from large DNA sequences. Bioinformatics 23 119–121. 10.1093/bioinformatics/btl519 [DOI] [PubMed] [Google Scholar]
- Amemura-Maekawa J., Kura F., Watanabe H. (1996). Cloning and nucleotide sequences of iron and copper-zinc superoxide dismutase genes of Legionella pneumophila and their distribution among Legionella species. Jpn. J. Med. Sci. Biol. 49 167–186. [DOI] [PubMed] [Google Scholar]
- Anderson K. L., Salyers A. A. (1989). Genetic evidence that outer membrane binding of starch is required for starch utilization by Bacteroides thetaiotaomicron. J. Bacteriol. 171 3199–3204. 10.1128/jb.171.6.3199-3204.1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arima K., Fujita H., Toita R., Imazu-Okada A., Tsutsumishita-Nakai N., Takeda N., et al. (2013). Amounts and compositional analysis of glycosaminoglycans in the tissue of fish. Carbohydr. Res. 366 25–32. 10.1016/j.carres.2012.11.010 [DOI] [PubMed] [Google Scholar]
- Avendaño-Herrera R., Magariños B., Moriñigo M., Romalde J. L., Toranzo A. E. (2005a). A novel O-serotype in Tenacibaculum maritimum strains isolated from cultured sole (Solea senegalensis). Bull. Eur. Assoc. Fish Pathol. 25 70–74. [Google Scholar]
- Avendaño-Herrera R., Núñez S., Barja J. L., Toranzo A. E. (2008). Evolution of drug resistance and minimum inhibitory concentration to enrofloxacin in Tenacibaculum maritimum strains isolated in fish farms. Aquac. Int. 16 1–11. 10.1007/s10499-007-9117-y [DOI] [Google Scholar]
- Avendaño-Herrera R., Toranzo A. E., Magariños B. (2006a). A challenge model for Tenacibaculum maritimum infection in turbot, Scophthalmus maximus (L.). J. Fish Dis. 29 371–374. 10.1111/j.1365-2761.2006.00712.x [DOI] [PubMed] [Google Scholar]
- Avendaño-Herrera R., Toranzo A. E., Magariños B. (2006b). Tenacibaculosis infection in marine fish caused by Tenacibaculum maritimum: a review. Dis. Aquat. Organ. 71 255–266. [DOI] [PubMed] [Google Scholar]
- Avendaño-Herrera R., Toranzo A. E., Romalde J. L., Lemos M. L., Magariños B. (2005b). Iron uptake mechanisms in the fish pathogen Tenacibaculum maritimum. Appl. Environ. Microbiol. 71 6947–6953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbeyron T., Thomas F., Barbe V., Teeling H., Schenowitz C., Dossat C., et al. (2016). Habitat and taxon as driving forces of carbohydrate catabolism in marine heterotrophic bacteria: example of the model algae-associated bacterium Zobellia galactanivorans Dsij(T). Environ. Microbiol. 18 4610–4627. 10.1111/1462-2920.13584 [DOI] [PubMed] [Google Scholar]
- Barriére C., Leroy-Sétrin S., Talon R. (2001). Characterization of catalase and superoxide dismutase in Staphylococcus carnosus 833 strain. J. Appl. Microbiol. 91 514–519. 10.1046/j.1365-2672.2001.01411.x [DOI] [PubMed] [Google Scholar]
- Baxa D. V., Kawai K., Kusuda R. (1988). In vitro and in vivo activities of Flexibacter maritimus toxins. Rep. United States Mar. Biol. Inst. Kochi Univ. 10 1–8. [Google Scholar]
- Bayliss S. C., Verner-Jeffreys D. W., Bartie K. L., Aanensen D. M., Sheppard S. K., Adams A., et al. (2017). The promise of whole genome pathogen sequencing for the molecular epidemiology of emerging aquaculture pathogens. Front. Microbiol. 8:121 10.3389/fmicb.2017.00121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardet J.-F., Campbell A. C., Buswell J. A. (1990). Flexibacter maritimus is the agent of “black patch necrosis” in Dover sole in Scotland. Dis. Aquat. Org. 8 233–237. 10.3354/dao008233 [DOI] [Google Scholar]
- Brown S. M., Howell M. L., Vasil M. L., Anderson A. J., Hassett D. J. (1995). Cloning and characterization of the katB gene of Pseudomonas aeruginosa encoding a hydrogen peroxide-inducible catalase: purification of KatB, cellular localization, and demonstration that it is essential for optimal resistance to hydrogen peroxide. J. Bacteriol. 177 6536–6544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burchard R. P., Rittschof D., Bonaventura J. (1990). Adhesion and motility of gliding bacteria on substrata with different surface free energies. Appl. Environ. Microbiol. 56 2529–2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva A. C., LePresle T., Capron A., Pierce R. J. (1992). Molecular cloning of a 16-kilodalton Cu/Zn superoxide dismutase from Schistosoma mansoni. Mol. Biochem. Parasitol. 52 275–278. 10.1016/0166-6851(92)90060-W [DOI] [PubMed] [Google Scholar]
- Dubin G. (2002). Extracellular proteases of Staphylococcus spp. Biol. Chem. 383 1075–1086. 10.1515/BC.2002.116 [DOI] [PubMed] [Google Scholar]
- Fillat M. F. (2014). The FUR (ferric uptake regulator) superfamily: diversity and versatility of key transcriptional regulators. Arch. Biochem. Biophys. 546 41–52. 10.1016/j.abb.2014.01.029 [DOI] [PubMed] [Google Scholar]
- Florio D., Gridelli S., Fioravanti M. L., Zanoni R. G. (2016). First isolation of Tenacibaculum maritimum in a captive sand tiger shark (Carcharias taurus). J. Zoo Wildl. Med. 47 351–353. 10.1638/2015-0064.1 [DOI] [PubMed] [Google Scholar]
- Fujita M. J., Kimura N., Yokose H., Otsuka M. (2012). Heterologous production of bisucaberin using a biosynthetic gene cluster cloned from a deep sea metagenome. Mol. Biosyst. 8 482–485. 10.1039/c1mb05431g [DOI] [PubMed] [Google Scholar]
- Fujita M. J., Nakano K., Sakai R. (2013). Bisucaberin B, a linear hydroxamate class siderophore from the marine bacterium Tenacibaculum mesophilum. Molecules 18 3917–3926. 10.3390/molecules18043917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg N., Manchanda G., Kumar A. (2014). Bacterial quorum sensing: circuits and applications. Antonie Van Leeuwenhoek 105 289–305. 10.1007/s10482-013-0082-3 [DOI] [PubMed] [Google Scholar]
- Gil R., Silva F. J., Peretó J., Moya A. (2004). Determination of the core of a minimal bacterial gene set. Microbiol. Mol. Biol. Rev. 68 518–537. 10.1128/MMBR.68.3.518-537.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert R. J. (2010). Cholesterol-dependent cytolysins. Adv. Exp. Med. Biol. 677 56–66. 10.1007/978-1-4419-6327-7_5 [DOI] [PubMed] [Google Scholar]
- Groisillier A., Hervé C., Jeudy A., Rebuffet E., Pluchon P. F., Chevolot Y., et al. (2010). MARINE-EXPRESS: taking advantage of high throughput cloning and expression strategies for the post-genomic analysis of marine organisms. Microb. Cell Fact. 9:45 10.1186/1475-2859-9-45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grothusen H., Castillo A., Henríquez P., Navas E., Bohle H., Araya C., et al. (2016). First complete genome sequence of Tenacibaculum dicentrarchi, an emerging bacterial pathogen of salmonids. Genome Announc. 4: e01756–15. 10.1128/genomeA.01756-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habib C., Houel A., Lunazzi A., Bernardet J. F., Olsen A. B., Nilsen H., et al. (2014). Multilocus sequence analysis of the marine bacterial genus Tenacibaculum suggests parallel evolution of fish pathogenicity and endemic colonization of aquaculture systems. Appl. Environ. Microbiol. 80 5503–5514. 10.1128/AEM.01177-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handlinger J., Soltani M., Percival S. (1997). The pathology of Flexibacter maritimus in aquaculture species in Tasmania, Australia. J. Fish Dis. 20 159–168. 10.1046/j.1365-2761.1997.00288.x [DOI] [Google Scholar]
- Ito M., Okino N., Tani M. (2014). New insight into the structure, reaction mechanism, and biological functions of neutral ceramidase. Biochim. Biophys. Acta 1841 682–691. 10.1016/j.bbalip.2013.09.008 [DOI] [PubMed] [Google Scholar]
- Kalia V. C. (2013). Quorum sensing inhibitors: an overview. Biotechnol. Adv. 31 224–245. 10.1016/j.biotechadv.2012.10.004 [DOI] [PubMed] [Google Scholar]
- Lau C. K., Krewulak K. D., Vogel H. J. (2016). Bacterial ferrous iron transport: the Feo system. FEMS Microbiol. Rev. 40 273–298. 10.1093/femsre/fuv049 [DOI] [PubMed] [Google Scholar]
- Lee M., Jun S. Y., Yoon B. Y., Song S., Lee K., Ha N. C. (2012). Membrane fusion proteins of type I secretion system and tripartite efflux pumps share a binding motif for TolC in gram-negative bacteria. PLoS ONE 7:e40460 10.1371/journal.pone.0040460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis A. L., Lewis W. G. (2012). Host sialoglycans and bacterial sialidases: a mucosal perspective. Cell Microbiol. 14 1174–1182. 10.1111/j.1462-5822.2012.01807.x [DOI] [PubMed] [Google Scholar]
- Li N., Qin T., Zhang X. L., Huang B., Liu Z. X., Xie H. X., et al. (2015). Gene deletion strategy to examine the involvement of the two chondroitin lyases in Flavobacterium columnare virulence. Appl. Environ. Microbiol. 81 7394–7402. 10.1128/AEM.01586-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López J. R., Núñez S., Magariños B., Castro N., Navas J. I., de la Herran R., et al. (2009). First isolation of Tenacibaculum maritimum from wedge sole, Dicologoglossa cuneata (Moreau). J. Fish Dis. 32 603–610. 10.1111/j.1365-2761.2009.01029.x [DOI] [PubMed] [Google Scholar]
- Los F. C., Randis T. M., Aroian R. V., Ratner A. J. (2013). Role of pore-forming toxins in bacterial infectious diseases. Microbiol. Mol. Biol. Rev. 77 173–207. 10.1128/MMBR.00052-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lujan K. M., Eisen J. A., Coil D. A. (2016). Draft genome sequence of Tenacibaculum soleae UCD-KL19. Genome Announc. 4: e01120–16. 10.1128/genomeA.01120-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magariños B., Pazos F., Santos Y., Romalde J. L., Toranzo A. E. (1995). Response of Pasteurella piscicida and Flexibacter maritimus to skin mucus of marine fish. Dis. Aquat. Organ. 21 103–108. 10.3354/dao021103 [DOI] [Google Scholar]
- Mally M., Shin H., Paroz C., Landmann R., Cornelis G. R. (2008). Capnocytophaga canimorsus: a human pathogen feeding at the surface of epithelial cells and phagocytes. PLoS Pathog. 4: e1000164 10.1371/journal.ppat.1000164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer C., Romero M., Muras A., Otero A. (2015). Aii20J, a wide-spectrum thermostable N-acylhomoserine lactonase from the marine bacterium Tenacibaculum sp. 20J, can quench AHL-mediated acid resistance in Escherichia coli. Appl. Microbiol. Biotechnol. 99 9523–9539. 10.1007/s00253-015-6741-8 [DOI] [PubMed] [Google Scholar]
- McBride M. J., Nakane D. (2015). Flavobacterium gliding motility and the type IX secretion system. Curr. Opin. Microbiol. 28 72–77. 10.1016/j.mib.2015.07.016 [DOI] [PubMed] [Google Scholar]
- McBride M. J., Xie G., Martens E. C., Lapidus A., Henrissat B., Rhodes R. G., et al. (2009). Novel features of the polysaccharide-digesting gliding bacterium Flavobacterium johnsoniae as revealed by genome sequence analysis. Appl. Environ. Microbiol. 75 6864–6875. 10.1128/AEM.01495-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride M. J., Zhu Y. (2013). Gliding motility and Por secretion system genes are widespread among members of the phylum Bacteroidetes. J. Bacteriol. 195 270–278. 10.1128/JB.01962-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel G., Pojasek K., Li Y., Sulea T., Linhardt R. J., Raman R., et al. (2004). The structure of chondroitin B lyase complexed with glycosaminoglycan oligosaccharides unravels a calcium-dependent catalytic machinery. J. Biol. Chem. 279 32882–32896. 10.1074/jbc.M403421200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson D. C., Garbe J., Collin M. (2011). Cysteine proteinase SpeB from Streptococcus pyogenes - a potent modifier of immunologically important host and bacterial proteins. Biol. Chem. 392 1077–1088. 10.1515/BC.2011.208 [DOI] [PubMed] [Google Scholar]
- Nelson S. S., Bollampalli S., McBride M. J. (2008). SprB is a cell surface component of the Flavobacterium johnsoniae gliding motility machinery. J. Bacteriol. 190 2851–2857. 10.1128/JB.01904-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda M., Takahashi M., Matsuno T., Uoo K., Nagahama M., Sakurai J. (2010). Hemolysis induced by Bacillus cereus sphingomyelinase. Biochim. Biophys. Acta 1798 1073–1080. 10.1016/j.bbamem.2010.03.004 [DOI] [PubMed] [Google Scholar]
- Okino N., Ikeda R., Ito M. (2010). Expression, purification, and characterization of a recombinant neutral ceramidase from Mycobacterium tuberculosis. Biosci. Biotechnol. Biochem. 74 316–321. 10.1271/bbb.90645 [DOI] [PubMed] [Google Scholar]
- Olczak T., Sroka A., Potempa J., Olczak M. (2008). Porphyromonas gingivalis HmuY and HmuR: further characterization of a novel mechanism of heme utilization. Arch. Microbiol. 189 197–210. 10.1007/s00203-007-0309-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazos F. (1997). Flexibacter Maritimus: Estudio Fenotípico, Inmunológico y Molecular. Ph.D. thesis, University of Santiago de Compostela, Santiago. [Google Scholar]
- Pers C., Gahrn-Hansen B., Frederiksen W. (1996). Capnocytophaga canimorsus septicemia in Denmark, 1982–1995: review of 39 cases. Clin. Infect. Dis. 23 71–75. 10.1093/clinids/23.1.71 [DOI] [PubMed] [Google Scholar]
- Rahman T., Suga K., Kinya K., Yukitaka S. (2014). Biological and serological characterization of a non-gliding strain of Tenacibaculum maritimum isolated from a diseased puffer fish Takifugu rubripes. Fish Pathol. 49 121–129. 10.3147/jsfp.49.121 [DOI] [Google Scholar]
- Ratledge C., Dover L. G. (2000). Iron metabolism in pathogenic bacteria. Annu. Rev. Microbiol. 54 881–941. 10.1146/annurev.micro.54.1.881 [DOI] [PubMed] [Google Scholar]
- Renzi F., Manfredi P., Mally M., Moes S., Jenö P., Cornelis G. R. (2011). The N-glycan glycoprotein deglycosylation complex (Gpd) from Capnocytophaga canimorsus deglycosylates human IgG. PLoS Pathog. 7:e1002118 10.1371/journal.ppat.1002118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero M., Avendaño-Herrera R., Magariños B., Cámara M., Otero A. (2010). Acylhomoserine lactone production and degradation by the fish pathogen Tenacibaculum maritimum, a member of the Cytophaga-Flavobacterium-Bacteroides (CFB) group. FEMS Microbiol. Lett. 304 131–139. 10.1111/j.1574-6968.2009.01889.x [DOI] [PubMed] [Google Scholar]
- Sasagawa Y., Izaki K., Matsubara Y., Suzuki K., Kojima H., Kamio Y. (1995). Molecular cloning and sequence analysis of the gene encoding the collagenase from Cytophaga sp. L43-1 strain. Biosci. Biotechnol. Biochem. 59 2068–2073. 10.1271/bbb.59.2068 [DOI] [PubMed] [Google Scholar]
- Sato K., Naito M., Yukitake H., Hirakawa H., Shoji M., McBride M. J., et al. (2010). A protein secretion system linked to bacteroidete gliding motility and pathogenesis. Proc. Natl. Acad. Sci. U.S.A. 107 276–281. 10.1073/pnas.0912010107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K., Yukitake H., Narita Y., Shoji M., Naito M., Nakayama K. (2013). Identification of Porphyromonas gingivalis proteins secreted by the Por secretion system. FEMS Microbiol. Lett. 338 68–76. 10.1111/1574-6968.12028 [DOI] [PubMed] [Google Scholar]
- Sheng Y., Abreu I. A., Cabelli D. E., Maroney M. J., Miller A. F., Teixeira M., et al. (2014). Superoxide dismutases and superoxide reductases. Chem. Rev. 114 3854–3918. 10.1021/cr4005296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R., Wiseman B., Deemagarn T., Jha V., Switala J., Loewen P. C. (2008). Comparative study of catalase-peroxidases (KatGs). Arch. Biochem. Biophys. 471 207–214. 10.1016/j.abb.2007.12.008 [DOI] [PubMed] [Google Scholar]
- Småge S. B., Frisch K., Brevik ØJ., Kuninori W., Nylunda A. (2016). First isolation, identification and characterisation of Tenacibaculum maritimum in Norway, isolated from diseased farmed sea lice cleaner fish Cyclopterus lumpus L. Aquaculture 464 178–184. 10.1016/j.aquaculture.2016.06.030 [DOI] [Google Scholar]
- Studier F. W. (2005). Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 41 207–234. 10.1016/j.pep.2005.01.016 [DOI] [PubMed] [Google Scholar]
- Suomalainen L. R., Tiirola M., Valtonen E. T. (2006). Chondroitin AC lyase activity is related to virulence of fish pathogenic Flavobacterium columnare. J. Fish Dis. 29 757–763. 10.1111/j.1365-2761.2006.00771.x [DOI] [PubMed] [Google Scholar]
- Suzuki M., Nakagawa Y., Harayama S., Yamamoto S. (2001). Phylogenetic analysis and taxonomic study of marine Cytophaga-like bacteria: proposal for Tenacibaculum gen. nov. with Tenacibaculum maritimum comb. nov. and Tenacibaculum ovolyticum comb. nov., and description of Tenacibaculum mesophilum sp. nov. and Tenacibaculum amylolyticum sp. nov. Int. J. Syst. Evol. Microbiol. 51 1639–1652. 10.1099/00207713-51-5-1639 [DOI] [PubMed] [Google Scholar]
- Teramoto M., Zhai Z., Komatsu A., Shibayama K., Suzuki M. (2016). Genome sequence of the psychrophilic bacterium Tenacibaculum ovolyticum strain da5A-8 isolated from deep seawater. Genome Announc. 4: e00644–16. 10.1128/genomeA.00644-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terrapon N., Lombard V., Gilbert H. J., Henrissat B. (2015). Automatic prediction of polysaccharide utilization loci in Bacteroidetes species. Bioinformatics 31 647–655. 10.1093/bioinformatics/btu716 [DOI] [PubMed] [Google Scholar]
- Vallenet D., Belda E., Calteau A., Cruveiller S., Engelen S., Lajus A., et al. (2013). MicroScope–an integrated microbial resource for the curation and comparative analysis of genomic and metabolic data. Nucleic Acids Res. 41 D636–D647. 10.1093/nar/gks1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gelderen R., Carson J., Nowak B. (2009). Effect of extracellular products of Tenacibaculum maritimum in Atlantic salmon, Salmo salar L. J. Fish Dis. 32 727–731. 10.1111/j.1365-2761.2009.01032.x [DOI] [PubMed] [Google Scholar]
- Varki N. M., Varki A. (2007). Diversity in cell surface sialic acid presentations: implications for biology and disease. Lab. Invest. 87 851–857. 10.1038/labinvest.3700656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernikos G. S., Parkhill J. (2006). Interpolated variable order motifs for identification of horizontally acquired DNA: revisiting the Salmonella pathogenicity islands. Bioinformatics 22 2196–2203. 10.1093/bioinformatics/btl369 [DOI] [PubMed] [Google Scholar]
- Waack S., Keller O., Asper R., Brodag T., Damm C., Fricke W. F., et al. (2006). Score-based prediction of genomic islands in prokaryotic genomes using hidden Markov models. BMC Bioinformatics 7:142 10.1186/1471-2105-7-142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakabayashi H., Hikida M., Masumura K. (1986). Flexibacter maritimus sp. nov., a pathogen of marine fishes. Int. J. Syst. Evol. Microbiol. 36 396–398. 10.1099/00207713-36-3-396 [DOI] [Google Scholar]
- Wójtowicz H., Wojaczyński J., Olczak M., Króliczewski J., Latos-Grażyński L., Olczak T. (2009). Heme environment in HmuY, the heme-binding protein of Porphyromonas gingivalis. Biochem. Biophys. Res. Commun. 383 178–182. 10.1016/j.bbrc.2009.03.148 [DOI] [PubMed] [Google Scholar]
- Xu Q., Rawlings N. D., Farr C. L., Chiu H. J., Grant J. C., Jaroszewski L., et al. (2011). Structural and sequence analysis of imelysin-like proteins implicated in bacterial iron uptake. PLoS ONE 6:e21875 10.1371/journal.pone.0021875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y., Mao X., Yang J., Chen X., Mao F., Xu Y. (2012). dbCAN: a web resource for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 40 W445–W451. 10.1093/nar/gks479 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.