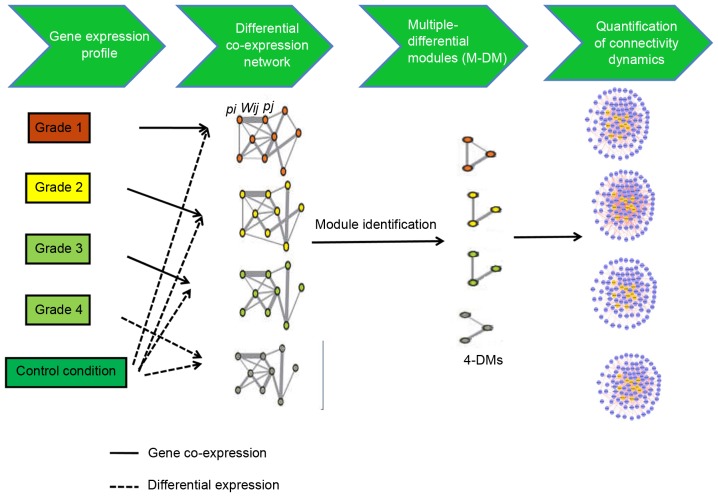
Figure 1.
Summary of the inference of multiple differential modules approach. This approach includes two main steps. In the first step, differential gene co-expression networks are constructed using microarray data across multiple conditions. To construct differential gene co-expression networks, a binary co-expression network is built where edges are selected according to the absolute value of Pearson correlation coefficient of the expression profiles of two genes. Only edges with correlation higher than the pre-defined threshold δ are chosen in the co-expression network. Then, edges in the binary co-expression network are weighted (Wx,y) on the basis of the P-values (Px and Py) of differential gene expression between control and disease conditions. During the second step, multiple differential gene co-expression networks are analyzed to extract shared multiple differential modules across different conditions. multiple differential modules with M=4 are the modules that are screened out under 4 conditions (grade 1, 2, 3, and 4 osteosarcoma).