Table 1. Data collection and refinement statistics for PSMA.
| Space group | P21 |
|---|---|
| Unit cell (a, b, c, β) | (74.9 Å, 157.8 Å, 133.8 Å, 93.2°) |
| Molecules per asymmetric unit | Two dimers |
| Resolution (outer shell) | 30-3.5 Å (3.62-3.5) |
| Theoretical reflections in resolution range | 77,001 |
| Working set | 70,431 (91.5%) |
| Test set | 3,671 (4.8%) |
| Completeness (outer shell)* | 99% (98.1) |
| Rmerge (outer shell)† | 14% (43.2) |
| I/σ (outer shell) | 15.4 (5.9) |
| Rcryst‡, Rfree§ | 25.2%, 28.4% |
| Average B factor (main chain, side chain) | 66 Å2 (54, 72) |
| non-H atoms per asymmetric unit | 22,876 |
| Protein, Zn(II), carbohydrate, water | 22, 184; 8; 672; 12 |
| rms deviation from ideality: bond, angle | 0.009 Å, 1.47° |
| Non-Gly residues in Ramachandran plot | |
| Most-favored | 74.2% |
| Additionally allowed | 22.9% |
| Generously allowed | 2% |
| Disallowed | 0.8% |
Completeness indicates number of independent reflections/total theoretical number.
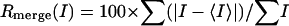 where I is the integrated intensity of a given reflection.
where I is the integrated intensity of a given reflection.
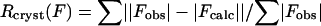 where |Fobs| and |Fcalc| are the observed and calculated structure-factor amplitudes for the HKL reflection.
where |Fobs| and |Fcalc| are the observed and calculated structure-factor amplitudes for the HKL reflection.
Rfree is calculated from reflections in a test set not included in the atomic refinement.
