Abstract
Pain is one of the most prominent symptoms of osteoarthritis. However, there is often discordance between the pain experienced by individuals with osteoarthritis and the degree of articular pathology. This suggests that individual differences, including genetic variability in the central processing of nociceptive stimuli, may impact the presentation of osteoarthritis. Here, we show that the single nucleotide polymorphism rs16868943 in the collagen gene COL11A2 is significantly associated with lowered heat pain tolerance on the arm in participants with knee osteoarthritis (P = 1.21 × 10−6, P = 0.0053 after Bonferroni correction, beta = −3.42). A total of 161 knee osteoarthritis participants were included and evaluated for heat, punctate and pressure pain sensitivity of the affected knee and the ipsilateral arm. Each participant was genotyped for 4392 single nucleotide polymorphisms in genes implicated in pain perception, inflammation and mood and tested for association with pain sensitivity. The minor A allele of single nucleotide polymorphism rs16868943 was significantly associated with lower arm heat pain tolerance after correction for age, gender, race, and study site. This single nucleotide polymorphism was also nominally associated with other measures of heat pain sensitivity, including lowered knee heat pain tolerance (P = 1.14 × 10−5, P = 0.05 after Bonferroni correction), lowered arm heat pain threshold (P = 0.0039, uncorrected) and lowered knee heat pain threshold (P = 0.003, uncorrected). Addition of genotypes from 91 participants without knee pain produced a significant interaction between knee osteoarthritis status and the rs16868943 single nucleotide polymorphism in heat pain tolerance (P = 1.71 × 10−5), such that rs16868943 was not associated with heat pain tolerance in participants without knee pain (P = 0.12, beta = 1.3). This is the first study to show genetic association with heat pain tolerance in individuals with osteoarthritis. The association is specific to participants who have already developed knee osteoarthritis, suggesting that the COL11A2 gene, which has previously been associated with familial osteoarthritis, may play a role in pain sensitization after the development of osteoarthritis.
Keywords: Osteoarthritis, knee osteoarthritis, pain genetics, association study, pain sensitivity, allodynia, COL11A2, heat pain, pain tolerance
Introduction
Pain is the most common symptom of osteoarthritis (OA), yet the determinants of OA-related pain remain poorly understood. Objective measures of OA severity are at best only modestly associated with pain severity, suggesting that individual differences in pain processing may contribute to OA-related pain. Indeed, heightened experimental pain sensitivity has been observed in participants with knee OA, and experimental pain sensitivity predicts severity of clinical pain in these participants.1–6 Understanding the factors driving heightened pain sensitivity in knee OA could provide clinically relevant information.
The experience of pain is a complex process determined by multiple biological and environmental factors, including genetic influences. The genetic contribution to pain sensitivity has been estimated to be 22–55% in twin studies.7 Heritability for nociceptive and analgesic sensitivity in mice similarly ranges from 28% to 76%.8 Several genes have been implicated in increased pain sensitivity in humans. The most-studied gene in pain research is the catechol-O-methyl-transferase (COMT) gene. COMT polymorphisms and haplotypes have been associated with both experimental pain sensitivity and clinical pain phenotypes. Several other genes have likewise shown associations with experimental pain sensitivity in multiple studies, including OPRM1,9 GCH1,10,11 MC1R,12 and SCN9A13,14 and dopamine transporter genes (SLC6A3 and SLC6A4).15
In OA-related pain, several genes have been associated with clinical pain severity in candidate gene studies. For example, the Val158Met polymorphism in the COMT gene (rs4680) was associated with increased hip OA pain,16 though a subsequent study failed to observe any association of this single nucleotide polymorphism (SNP) with knee OA.17 Also, SNP rs900414 in the PCSK6 gene was associated with protection against pain when radiographic OA was present.18
Despite these recent advances in our knowledge of genetic contributions to OA, our understanding of genetic factors associated with pain sensitivity in OA remains limited. Previous studies have focused on a few very specific variants in candidate genes, making it difficult to discover potential disease-associated variants outside of the hypotheses. The primary aim of this study is to significantly expand the tested genes and polymorphisms, with the hope to discover new risk genes and alleles for pain sensitivity in OA. To address this aim, we performed an association study including 4392 SNPs in 555 genes implicated in pain, inflammation, and psychiatric disorders. We performed extensive pain phenotyping, including assessment of clinical pain and quantitative sensory testing to assess sensitivity to heat, cold, pressure, and punctate pain. To characterize pain sensitivity at sites remote from the primary painful joint in OA participant, pain sensitivities were assessed not only on the most painful knee (i.e., index knee) but also at body sites outside the index knee.
Material and methods
Study subjects
Recruitment of knee OA participants in the study has been described elsewhere.19 Briefly, 45- to 85-year-old individuals, who self-identified their race/ethnicity as African American or Non-Hispanic Whites, were enrolled in the study at the University of Florida and University of Alabama at Birmingham. Participants were recruited who screened positive for unilateral or bilateral symptomatic knee OA20 and physical examination subsequently confirmed knee pain symptoms consistent with a clinical diagnosis of knee OA, regardless of radiographic evidence. At the time of study entry, posteroanterior and lateral radiographs of the knees were obtained from all participants, with the knees in a bilateral weight-bearing, fixed-flexion position, as described elsewhere.21
Participants were excluded if any of the following features were present: (1) prosthetic knee replacement or other clinically significant surgery to the affected knee; (2) uncontrolled hypertension (blood pressure >150/95 mm Hg), heart failure, or history of acute myocardial infarction; (3) peripheral neuropathy; (4) systemic rheumatic disorders including rheumatoid arthritis, systemic lupus erythematosus, and fibromyalgia; (5) daily opioid use; (6) cognitive impairment; or (7) hospitalization for psychiatric illness within the preceding year. Individuals with no knee pain (i.e., controls) met the same inclusion/exclusion criteria, with the exception that they reported no knee pain.
A total of 177 participants with knee OA and 99 participants without knee pain were genotyped. Participants with missing gender, age, race, or study sites were excluded from the analysis. Participants with greater than 10% missing genotype were also excluded. After exclusion criteria were applied, 161 knee OA and 91 participants without knee pain were included in the analysis.
Pain measurement
Pain measurements have been described elsewhere.19 The index knee here is defined as the most painful knee.
Heat
Heat–pain threshold and tolerance levels were assessed on both the index knee and the ipsilateral ventral forearm, starting at the baseline temperature (32℃) and increasing 0.5℃/s. Participants were instructed to press a button when the sensation “first became painful” (i.e., threshold) and also when they “no longer felt able to tolerate the pain” (i.e., tolerance). The mean temperature from three trials was used for analysis.
Pressure
Pressure–pain thresholds were evaluated at the medial joint line of the index knee as well as the ipsilateral trapezius. For each site, a handheld Medoc digital pressure Algometer (Algomed) was applied at a constant rate of 30 kPa/s. The participant was instructed to press a button when the sensation “first became painful.” An average pressure–pain threshold was determined for each site from three trials. The maximum pressure for the knee site was 600 and 1000 kPa at the ipsilateral trapeziius. If participants did not report pain at the maximum pressure level, the procedure was terminated and a pressure of 600 or 1000 kPa was assigned for that trial.
Punctate
Sensitivity to punctate mechanical stimuli was assessed at the index patella and back of the ipsilateral hand using a nylon monofilament delivering a target force of 300 g. Participants provided verbal pain ratings following a single contact and after 10 contacts, at a rate of 1 contact/s. Ratings were made on a scale of 0 to 100. The procedure was repeated, and the ratings for multiple contacts were averaged for analysis.
Cold sensitivity
Participants placed their right hand into a 12℃ cold water bath for up to 1 min. Participants provided verbal ratings of pain intensity at the end of the immersion on a scale of 0 to 100. If a participant withdrew the hand from the water bath before the 1-min period ended, the pain rating was collected and the withdrawal time was recorded.
Graded Chronic Pain Scale
The Graded Chronic Pain Scale (GCPS) evaluates global pain severity and pain-related interference over the past six months and consists of seven items related to pain intensity and pain interference.22 With a 0–10 numeric rating scale, participants rated the intensity of their current knee pain and the worst and average pain during the past six months. These three items were averaged and multiplied by 10 to generate a GCPS characteristic pain intensity score.
Western Ontario and McMaster Universities Osteoarthritis Index
The Western Ontario and McMaster Universities Osteoarthritis Index (WOMAC)23 assesses symptoms of knee OA in the past 48 h. For this study, the 4-point Likert-type scale version was used. The WOMAC yields three subscales, including pain during activities (5 items), stiffness during the day (2 items), and impairments in physical function (17 items), with higher scores indicating worse pain, stiffness, and impairments in physical function.
Genotyping
Knee OA participants and healthy participants were genotyped in the same fashion. At enrollment, whole blood was collected by venipuncture from study participants who provided consent for genotyping. Blood was collected into 5-mL ethylenediamine tetra-acetic acid–containing Vacutainer tubes (Beckton Dickinson, Franklin Lakes, NJ), and leukocytes were purified and stored at −80℃. Genomic DNA was purified utilizing protocols based on PureGene Extraction Kits from Qiagen (Germantown, MD). All individuals were genotyped using the Algynomics (Chapel Hill, NC) Pain Research Panel, a dedicated chip-based platform manufactured by Illumina. The panel assesses 4900 SNPs representing 555 genes known to be involved in systems relevant to pain perception (complete list provided in the Supplementary Material 1). DNA sample concentrations were measured with PicoGreen (ThermoFisher Scientific, Waltham, MA) and diluted to 50 ng/µl in 96-well plates. These were genotyped using the Algynomics chips, run on an Illumina VeraCode instrument (Illumina, San Diego, CA). SNPs were excluded when minor allele frequency was 1% or less, Hardy–Weinberg equilibrium P < 10−5, and SNP call rate 95% or less. After quality control, a total of 4392 SNPs were included in the analysis.
Statistical analysis
Associations between each phenotype and each individual SNP were tested with PLINK using the additive genetic model in linear regression,24 with age, sex, study sites, and race as covariates. A similar analysis including weight as an additional covariate was applied to the significantly associated SNP. Bonferroni correction for testing 4392 SNPs was applied to the adjusted P value. The study-wide Bonferroni-corrected statistical significance level was set at P = 1.14 × 10−5. A subanalysis was carried out in Africans Americans and Europeans Americans for heat pain tolerance and threshold. The rs16868943 polymorphism, being the only significant SNP associated with the arm heat tolerance phenotype after Bonferroni correction, was selected for further analysis in participants without knee pain. In this population, a similar association study was carried out between rs16868943 and arm heat tolerance. A subanalysis was carried out in Africans and European Americans. The interaction between the heat pain tolerance in the arm and knee OA phenotype was also tested. Interaction P value was calculated using the linear regression model in PLINK, controlling for age, sex, study sites, and race. The locus plot was generated by LocusZoom.25 Comparisons of pain measures between knee OA and participants without knee pain were calculated with two-tailed Student’s t-test.
Results
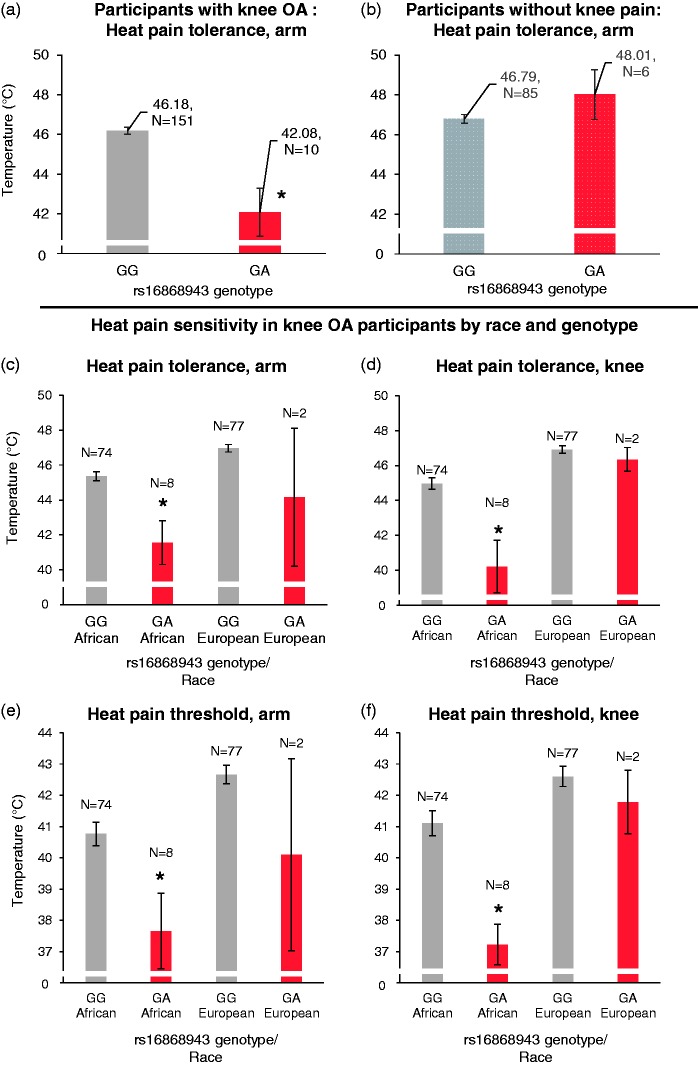
A total of 161 participants with knee OA and 4392 SNPs in 555 genes were included in the final analysis after quality control. The average age of the participants was 56.8 years, with 73% female and 27% male, 49% European Americans, and 51% African Americans. Ninety-one participants without knee pain were also included for comparison, with an average age of 56.8 years, 32% male and 68% female, 75% European Americans, and 25% African Americans. Participants with knee OA, compared to participants without knee pain, in general had elevated sensitivity to heat, pressure, and punctate pain in both the index knee and ipsilateral arm (Table 1). This is consistent with previous findings from this cohort and others showing that participants with knee OA have generalized pain sensitization.23,26–29 Association studies were carried out between SNPs and clinical as well as experimental pain measures with linear regression, adjusting for age, gender, race, and study sites. The top associated SNPs for each pain measure are listed in Table 2. Based on the study-wide Bonferroni-corrected statistical significance level of P = 1.14 × 10−5, the only SNP that reached statistical significance for association with any pain measure was rs16868943 in the COL11A2 gene, which was found to be associated with lower heat pain tolerance in the ipsilateral arm at P = 1.21 × 10−6 (P = 0.0053 after Bonferroni correction, and beta = −3.42). Because weight is a risk factor for knee OA pain, we added weight as an additional covariate for comparison but it did not significantly alter the association P value (P = 1.08 × 10−6). The average heat pain tolerance in knee OA participants with the GG genotype is 46.18℃, but the tolerance for participants with the GA genotype is 42.08℃ (Figure 1(a)). There was no AA genotype present in our data set, likely secondary to the low minor allele frequency. This SNP was also marginally associated with lower heat pain tolerance in the index knee (P = 1.14 × 10−5, corrected P = 0.05, beta = −3.67), lower heat pain threshold in the ipsilateral arm (P = 0.0039, corrected P = 1, beta = −2.83) and knee (P = 0.0030, corrected P = 1, beta = −2.9), higher punctate pain sensitivity in the ipsilateral hand (P = 0.0065, corrected P = 1, beta = 22) and higher cold pain sensitivity (P = 0.049, corrected P = 1, beta = 21.58). It was not associated with punctate pain in the index knee (P = 0.30), pressure pain of the index knee (P = 0.39), or ipsilateral trapezius (P = 0.32), WOMAC score (P = 0.33), or the GCPS score (P = 0.77) (Table 2).
Table 1.
Demographics and pain sensitivity of knee OA and healthy participants.
| Phenotype | Knee OA (N = 161) | No knee pain (N = 91) | P |
|---|---|---|---|
| Age | 56.83 | 56.81 | 0.99 |
| Female (%) | 73 | 68 | 0.34 |
| African American (%) | 51 | 25 | 0.000067 |
| Annual Income (10k) | 4.25 | 4.75 | 0.21 |
| Heat pain threshold, ipsilateral arm (℃) | 41.5 | 42.74 | 0.0017 |
| Heat pain threshold, index knee (℃) | 41.64 | 42.23 | 0.16 |
| Heat pain tolerance, ipsilateral arm (℃) | 45.92 | 46.87 | 0.0017 |
| Heat pain tolerance, index knee (℃) | 45.68 | 46.68 | 0.0013 |
| Pressure pain threshold, index knee (kPa) | 288.41 | 369.04 | 0.00014 |
| Pressure pain threshold, ipsilateral trapezius(kPa) | 271.54 | 350.24 | 0.0015 |
| Punctate pain, index knee (out of 100) | 34.68 | 23.81 | 0.0032 |
| Punctate pain, ipsilateral hand (out of 100) | 25.33 | 14.61 | 0.00034 |
Participants with knee OA in general have greater pain sensitivity compared to participants without knee pain. P value is calculated from two-tailed student’s t-test.
OA: osteoarthritis.
Table 2.
Top SNPs associated with each pain measure.
| SNP | Gene | Functional consequence | Chr | Position | Major allele | Minor allele | Minor allele frequency | Beta | P (uncorrected) | P (Bonferroni corrected) |
|---|---|---|---|---|---|---|---|---|---|---|
| Heat pain tolerance, ipsilateral arm | ||||||||||
| rs16868943 | COL11A2 | Intron variant | 6 | 33147727 | G | A | 0.034 | −3.42 | 1.21 × 10−6a | 0.0053a |
| rs11129802 | SCN10A | Intron variant | 3 | 38750436 | C | T | 0.17 | 1.06 | 0.00024 | 1 |
| Heat pain tolerance, index knee | ||||||||||
| rs16868943 | COL11A2 | Intron variant | 6 | 33147727 | G | A | 0.034 | −3.67 | 1.14 × 10−5 | 0.05 |
| Heat pain threshold, ipsilateral arm | ||||||||||
| rs2020933 | SLC6A4 | Intron variant | 17 | 28561755 | A | T | 0.15 | 1.59 | 0.0004 | 1 |
| rs16868943 | COL11A2 | Intron variant | 6 | 33147727 | G | A | 0.034 | −2.83 | 0.0039 | 1 |
| Heat pain threshold, index knee | ||||||||||
| rs7103411 | BDNF-AS, BDNF | Intron variant | 11 | 27700125 | T | C | 0.25 | 2.29 | 0.0001 | 0.44 |
| rs16868943 | COL11A2 | Intron variant | 6 | 33147727 | G | A | 0.034 | −2.90 | 0.0030 | 1 |
| Punctate-induced pain, ipsilateral hand | ||||||||||
| rs12415832 | GRK5 | Intron variant | 10 | 121112327 | C | A | 0.055 | 25.02 | 0.00046 | 1 |
| rs16868943 | COL11A2 | Intron variant | 6 | 33147727 | G | A | 0.034 | 22.00 | 0.0065 | 1 |
| Punctate-induced pain, index knee | ||||||||||
| rs2398144 | GNAO1 | Intron variant | 16 | 56352854 | A | C | 0.43 | −9.94 | 0.00051 | 1 |
| rs16868943 | COL11A2 | Intron variant | 6 | 33147727 | G | A | 0.034 | 8.78 | 0.30 | 1 |
| Pressure-induced pain threshold, index knee | ||||||||||
| rs3790112 | GNAO1 | Intron variant | 16 | 56381197 | G | T | 0.38 | −61.51 | 0.00013 | 0.57 |
| rs16868943 | COL11A2 | Intron variant | 6 | 33147727 | G | A | 0.034 | −41.10 | 0.39 | 1 |
| Pressure-induced pain threshold, ipsilateral trapezius | ||||||||||
| rs1059829 | SPARC | utr variant 3 prime | 5 | 151042029 | G | A | 0.39 | 65.96 | 0.00013 | 0.57 |
| rs16868943 | COL11A2 | Intron variant | 6 | 33147727 | G | A | 0.034 | −52.64 | 0.32 | 1 |
| Cold sensitivity | ||||||||||
| rs1479922 | IL2 | Intergenic | 4 | 123371785 | G | A | 0.016 | 36.05 | 0.00071 | 1 |
| rs16868943 | COL11A2 | Intron variant | 6 | 33147727 | G | A | 0.034 | 21.58 | 0.049 | 1 |
| Graded chronic pain scale | ||||||||||
| rs6812524 | SPP1 | Synonymous codon | 4 | 88902725 | G | A | 0.073 | 12.15 | 0.00072 | 1 |
| rs16868943 | COL11A2 | Intron variant | 6 | 33147727 | G | A | 0.034 | 2.10 | 0.77 | 1 |
| Western Ontario and McMaster Universities Osteoarthritis Index | ||||||||||
| rs17586428 | HTR2B, PSMD1 | Intron variant | 2 | 231988855 | A | G | 0.18 | 2.20 | 0.00033 | 1 |
| rs16868943 | COL11A2 | Intron variant | 6 | 33147727 | G | A | 0.034 | −1.34 | 0.33 | 1 |
The study-wide significant P value is set at 1.14 × 10−5 after Bonferroni correction for 4392 SNPs included in the study.
SNP: single nucleotide polymorphism.
rs16868943 is associated with decreased heat pain tolerance after Bonferroni correction. Associations of this SNP with each pain measure are included in this table.
Figure 1.
Heat pain sensitivity by phenotype and genotype. (a) The s16868943 polymorphism is associated with decreased arm heat pain tolerance in participants with knee OA (P = 1.21 × 10−6, P = 0.0053 after Bonferroni correction, beta = −3.42), but not in participants without knee pain; (b) the heat pain tolerance and threshold are broken down by race and genotype in (c–f). The GA genotype is nominally associated with lower heat pain tolerance and threshold in the index knee in African Americans but not in European Americans. *Statistically significant association of s16868943 with the specific phenotype (uncorrected P < 0.05). For a complete listing of uncorrected and corrected P values, please refer to the Results section.
Because the minor allele frequency (A allele) of this SNP is higher in African Americans than European Americans, associations with heat pain tolerance/threshold were examined separately by race. Knee OA participants with the GA genotype, compared to the GG genotype, had lower heat pain tolerance/threshold in the ipsilateral arm and ipsilateral leg within both African Americans and European Americans (Figure 1 (c)—(f). Within African Americans, the presence of the A allele was associated with lower heat pain tolerance in the index knee (P = 4.91 × 10−5, corrected P = 0.21, beta = −4.68) and the ipsilateral arm (P = 3.21 × 10−5, corrected P = 0.14, beta = −3.74), and lower heat pain threshold in the index knee (P = 0.0037, corrected P = 1, beta = −3.59) and ipsilateral arm (P = 0.0018, corrected P = 1, beta = −2.90), compared to the G allele. Within European Americans, the A allele was not statistically associated with heat pain tolerance in the index knee (P = 0.91, beta = −0.14), the ipsilateral arm (P = 0.072, beta = −2.38), or heat pain threshold in the index knee (P = 0.77, beta = −2.57) and ipsilateral arm (P = 0.25, beta = −2.09). The P value approached significance for the ipsilateral arm in European Americans, but there was reduced power due to the lower A allele frequency in this population. The direction of the association was the same between European and African Americans, with presence of the A allele related to reduced pain tolerance.
We carried out a follow-up association study between rs16868943 and participants without knee pain, to test whether this polymorphism also is associated with decreased heat pain tolerance in participants without knee pain. The result is shown in Figure 1(b). There was no association found between arm heat pain tolerance and rs16868943 in participants without knee pain (P = 0.12). The data trend was also opposite of that seen in participants with knee OA (beta = 1.3). A subanalysis was carried out in African Americans and European Americans without knee pain (Supplementary Material 2, Figure 1). In African Americans, there was no association between arm heat pain tolerance and the minor allele of rs16868943 (P = 0.31, beta = −0.85). In European Americans, there was a significant association (P = 0.03), but the trend was opposite of that seen in participants with knee OA (beta = 2.68). Because of the discrepancy seen between participants with knee OA and participants without knee pain, we asked whether the genotypes of rs16868943 interact with the presence of knee OA. After controlling for race, gender, age, and study site, a significant interaction was found between the minor allele of rs16868943 and the presence of knee OA (P = 1.71 × 10−5, see Supplementary Material 2, Figure 2 for illustration.). This result suggests that the effect of this polymorphism is dependent upon the development of knee OA. Its effect on pain perception is not present in participants without knee pain.
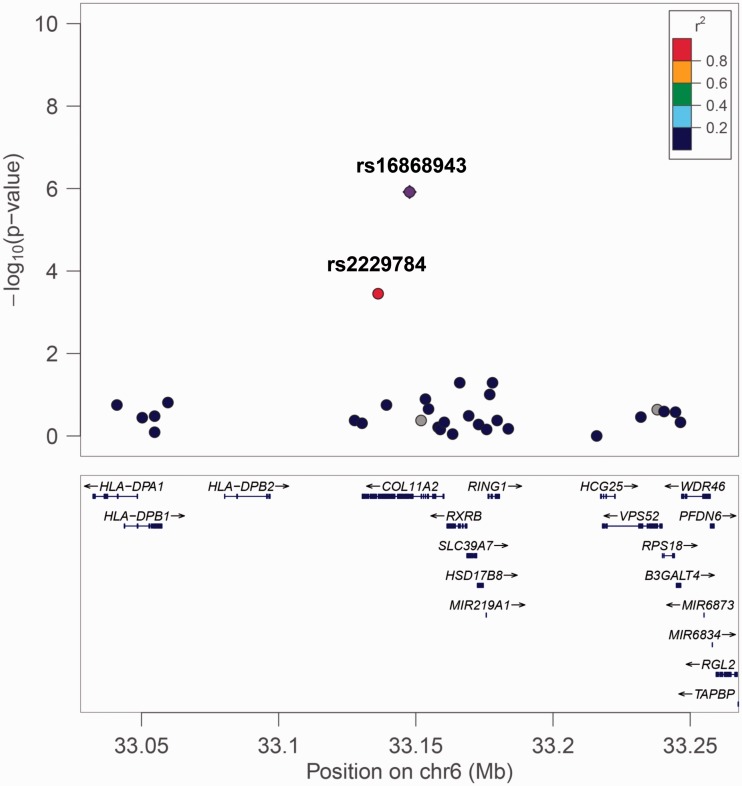
The genomic structure and P values of SNPs surrounding rs16868943 are shown in Figure 2. The rs16868943 polymorphism is in high linkage disequilibrium (LD) with many local SNPs, but only rs2229784 is included in the study. The rs2229784 polymorphism is a missense mutation (Pro → Thr) variant within the COL11A2 gene. It is associated with lower arm heat pain tolerance at P = 0.00035 (corrected P = 1). This finding implies that the rs16868943 polymorphism, an intron variant in the COL11A2 gene, and the rs2229784 polymorphism, a missense variant in the COL11A2 gene may both play a role in the development of increased pain sensitivity, or that other genetic variants within the haplotype are conferring or contributing to the effect.
Figure 2.
Genetic structure around the s16868943 polymorphism. The rs16868943 polymorphism lies in the COL11A2 gene. The rs2229784 polymorphism is a missense mutation (Pro →Thr) in the COL11A2 gene in strong linkage disequilibrium with rs16868943.
Discussion
Pain sensitivity is influenced by multiple individual difference factors, including genetic variability. We hypothesize that natural genetic variations within pain-related genes affect the development of such hypersensitivity in participants with knee OA. Here, we showed that the minor (A) allele of rs16868943 in COL11A2 is significantly associated with lowered arm heat tolerance in participants with knee OA. This SNP is also nominally associated with other measures of heat pain hypersensitivity, including lowered knee heat pain tolerance, lowered arm heat pain threshold, and lowered knee heat pain threshold. Since minor allele frequency is higher in the African American population (7%) compared to the non-Hispanic Whites (1.4%), a race-specific analysis was carried out. The same finding remains significant in African Americans and the trend is similar within non-Hispanic Whites. Furthermore, an association study carried out between rs16868943 and arm heat pain tolerance was found to be non-significant in participants without knee pain.
The rs16868943 SNP is located within an intron of the COL11A2 gene, coding for the protein Collagen Type XI Alpha 2 Chain. This SNP is scored a 5 (TF binding or DNase peak) on RegulomeDB,30 with unclear functional implication. However, this SNP is in high LD with several SNPs with high regulatory potentials. Over 20 SNPs are in high LD (R2 > 0.7), with these SNPs in the European American and African American population (for details, please see LDProxy using the ASW and CEU population on the LDlink website31). These SNPs may represent potential causative SNPs. Most of these SNPs are within the COL11A2 gene, including the missense variant rs2229784, but some of these SNPs are within other nearby genes, including HLA-DPA1, HLA-DPB1, HLA-DPB2, and RING1. Therefore, rs16868943 may simply be a proxy for other causative, genetically linked polymorphisms. The A allele of rs16868943 has also been associated with Celiac disease in a GWAS study32 at a genome-wide significant level (P = 2.06 × 10−11). The clinical significance of such an association is unclear.
The COL11A2 gene has been linked to the development of OA in several studies. A splicing mutation in the gene has been associated with early onset OA.33 Mutations in COL11A2 have also been associated with Stickler’s syndrome,34,35 a syndrome commonly accompanied with joint pain, stiffness, and inflammation. It is also the disease gene for the autosomal-dominant and recessive forms of osteochondrodysplasia.36 In animal model, the COL11A2 knock-out mouse shows an increased rate of developing knee OA.37 COL11A2 has not been associated with OA in several recent GWAS studies,38–40 but its sister gene, COL11A1, has been associated with OA in a meta-analysis of nine genome-wide association studies.41 Despite these findings, no study to date has suggested the role of COL11A2 in pain perception among individuals with OA. This is the first study to suggest that the A allele of rs16868943, within COL11A2, is associated with increased heat pain sensitivity in participants with knee OA. This finding expands the potential influence of the COL11A2 gene to not just the development of OA but potentially to the development of the widespread painful sensation in participants with OA.
The minor allele frequency of rs16868943 differs among racial populations, being higher in the African American population (7%) than the European Americans (1.4%). African Americans in general have enhanced sensitivity to noxious stimuli, including lower heat pain tolerance compared to their European American counterparts.19,42–44 The higher allele frequency of this polymorphism in African Americans may partially contribute to their lower pain tolerance compared to European Americans. Despite the higher allele frequency, our analysis showed that African Americans still displayed higher pain sensitivity and clinical pain levels after controlling for the rs16868943 genotype. This finding suggests that there are factors other than the rs16868943 genotype causing the lower pain tolerance within African Americans, whether it be other genetic or environmental factors.
The mechanism by which COL11A2 gene produces heightened heat pain sensitivity in people with knee OA is unclear. One possibility is that COL11A2 is related to the pathology of OA and heightened pain sensitivity is the indirect consequence of the chronic OA disease. However, multiple GWAS studies have failed to show an association of COL11A2 with OA, which argues against COL11A2 as a causative OA gene. Perhaps the effect of COL11A2 is somehow activated by the genetic predispositions of those who developed knee OA. This is consistent with the significant interaction in heat pain sensitivity between the presence of knee OA and the genotype of rs16868943, suggesting that this polymorphism does not play a role in heat pain until knee OA has developed. Given that COL11A1, a sister gene of COL11A2, has been found to be associated with OA in a recent meta-analysis, one possible genetic explanation is that COL11A1 and COL11A2 may be working in concert in increasing heat pain sensitivity. Unfortunately, COL11A1 was not included in our study so this hypothesis could not be addressed. Another interesting observation is that COL11A2 solely affects heat pain sensitivity and thresholds but not other clinical or experimental pain measures. The reason behind such a difference is again unclear. COL11A2 may be only involved in the transduction pathway of heat pain45 but not pathways involving cold, punctuate, or pressure pain.
Central pain sensitization has been hypothesized as one of the mechanisms producing pain in individuals with OA. Quantitative sensory studies indicated that OA individuals are more sensitive to experimental pain stimuli than healthy individuals, particularly among individuals with high levels of clinical pain.26–29 Notably, the increased pain sensitivity is not only present at sites close to the affected knee29 but also at sites remote from the primary painful joint, including the forehead46 and the arms.29,47 Within our study, the rs16868943 polymorphism of COL11A2 conferred decreased heat pain tolerance and threshold in both the ipsilateral arm and knee. This indicates that the effect of COL11A2 on heat pain responses is widespread and not just limited to the ipsilateral knee. Such an observation suggests the possible role of COL11A2 in producing wide-spread pain sensitization in knee OA.
There are several limitations to this study. First, the findings are limited to the genes and SNPs included in this study. Second, all the experimental tasks were acute, controlled painful experiences. Given the artificial nature of the experimental procedures, the outcomes may have limited practical utility. However, several studies have shown the relevance of using experimental pain induction procedures to predict clinical pain1–6 In addition, our sample size was relatively small for a candidate gene study, especially given the low minor allele frequency of rs16868943 in non-Hispanic whites. Thus, replication of these findings in a larger sample is needed. Another limitation is that because it is a retrospective study, it is difficult to know whether the participants developed higher pain sensitivities after developing knee OA. A prospective study is needed to more confidently conclude that the rs16868943 A allele, or related haplotype, is associated with increased pain sensitivity before and after onset of knee OA. Because this study was initially designed to examine ethnic differences, roughly half of the participants were African American and half were European Americans. Although self-reported ethnicity was controlled in the linear regression analysis, underlying population stratification may have confounded the outcome.
These limitations notwithstanding, our findings indicate that the rs16868943 polymorphism in the COL11A2 gene may be a marker for increased heat pain sensitivity in participants with knee OA. Additional research is warranted to replicate and expand this finding.
Declaration of Conflicting Interests
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding
The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This research was supported by NIH/NIA Grant Numbers R37AG033906 (awarded to both the University of Florida and the University of Alabama at Birmingham) and K07AG046371 (RBF), and K99AG052642 (EJB). The research was also supported by NIH Clinical and Translational Science Awards UL1TR001417 (University of Alabama at Birmingham) and UL1TR001427 (University of Florida). Dr. Emily Bartley received support from the National Institutes of Health (K99AG052642). KTS is funded by the National Institute of Arthritis and Musculoskeletal and Skin Diseases, K23AR062099.
References
- 1.Langemark M, Jensen K, Jensen TS, et al. Pressure pain thresholds and thermal nociceptive thresholds in chronic tension-type headache. Pain 1989; 38: 203–210. [DOI] [PubMed] [Google Scholar]
- 2.Fillingim RB, Maixner W, Kincaid S, et al. Pain sensitivity in patients with temporomandibular disorders: relationship to clinical and psychosocial factors. Clin J Pain 1996; 12: 260–269. [DOI] [PubMed] [Google Scholar]
- 3.Clauw DJ, Williams D, Lauerman W, et al. Pain sensitivity as a correlate of clinical status in individuals with chronic low back pain. Spine 1999; 24: 2035–2041. [DOI] [PubMed] [Google Scholar]
- 4.Edwards RR, Doleys DM, Lowery D, et al. Pain tolerance as a predictor of outcome following multidisciplinary treatment for chronic pain: differential effects as a function of sex. Pain 2003; 106: 419–426. [DOI] [PubMed] [Google Scholar]
- 5.Edwards RR, Ness TJ, Weigent DA, et al. Individual differences in diffuse noxious inhibitory controls (DNIC): association with clinical variables. Pain 2003; 106: 427–437. [DOI] [PubMed] [Google Scholar]
- 6.Granot M, Lowenstein L, Yarnitsky D, et al. Postcesarean section pain prediction by preoperative experimental pain assessment. Anesthesiology 2003; 98: 1422–1426. [DOI] [PubMed] [Google Scholar]
- 7.Norbury TA, MacGregor AJ, Urwin J, et al. Heritability of responses to painful stimuli in women: a classical twin study. Brain 2007; 130: 3041–3049. [DOI] [PubMed] [Google Scholar]
- 8.Mogil JS. The genetic mediation of individual differences in sensitivity to pain and its inhibition. Proc Natl Acad Sci U S A 1999; 96: 7744–7751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fillingim RB, Kaplan L, Staud R, et al. The A118G single nucleotide polymorphism of the mu-opioid receptor gene (OPRM1) is associated with pressure pain sensitivity in humans. J Pain 2005; 6: 159–167. [DOI] [PubMed] [Google Scholar]
- 10.Tegeder I, Costigan M, Griffin RS, et al. GTP cyclohydrolase and tetrahydrobiopterin regulate pain sensitivity and persistence. Nat Med 2006; 12: 1269–1277. [DOI] [PubMed] [Google Scholar]
- 11.Tegeder I, Adolph J, Schmidt H, et al. Reduced hyperalgesia in homozygous carriers of a GTP cyclohydrolase 1 haplotype. Eur J Pain 2008; 12: 1069–1077. [DOI] [PubMed] [Google Scholar]
- 12.Mogil JS, Ritchie J, Smith SB, et al. Melanocortin-1 receptor gene variants affect pain and mu-opioid analgesia in mice and humans. J Med Genet 2005; 42: 583–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reimann F, Cox JJ, Belfer I, et al. Pain perception is altered by a nucleotide polymorphism in SCN9A. Proc Natl Acad Sci U S A 2010; 107: 5148–5153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Emery EC, Habib AM, Cox JJ, et al. Novel SCN9A mutations underlying extreme pain phenotypes: unexpected electrophysiological and clinical phenotype correlations. J Neurosci 2015; 35: 7674–7681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mogil JS. Pain genetics: past, present and future. Trends Genet 2012; 28: 258–266. [DOI] [PubMed] [Google Scholar]
- 16.van Meurs JB, Uitterlinden AG, Stolk L, et al. A functional polymorphism in the catechol-O-methyltransferase gene is associated with osteoarthritis-related pain. Arthritis Rheum 2009; 60: 628–629. [DOI] [PubMed] [Google Scholar]
- 17.Neogi T, Soni A, Doherty SA, et al. Contribution of the COMT Val158Met variant to symptomatic knee osteoarthritis. Ann Rheum Dis 2014; 73: 315–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Malfait AM, Seymour AB, Gao F, et al. A role for PACE4 in osteoarthritis pain: evidence from human genetic association and null mutant phenotype. Ann Rheum Dis 2012; 71: 1042–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cruz-Almeida Y, Sibille KT, Goodin BR, et al. Racial and ethnic differences in older adults with knee osteoarthritis. Arthritis Rheumatol 2014; 66: 1800–1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roux CH, Saraux A, Mazieres B, et al. Screening for hip and knee osteoarthritis in the general population: predictive value of a questionnaire and prevalence estimates. Ann Rheum Dis 2008; 67: 1406–1411. [DOI] [PubMed] [Google Scholar]
- 21.Felson DT, Nevitt MC, Yang M, et al. A new approach yields high rates of radiographic progression in knee osteoarthritis. J Rheumatol 2008; 35: 2047–2054. [PMC free article] [PubMed] [Google Scholar]
- 22.Von Korff M, Ormel J, Keefe FJ, et al. Grading the severity of chronic pain. Pain 1992; 50: 133–149. [DOI] [PubMed] [Google Scholar]
- 23.Collins NJ, Misra D, Felson DT, et al. Measures of knee function: International Knee Documentation Committee (IKDC) Subjective Knee Evaluation Form, Knee Injury and Osteoarthritis Outcome Score (KOOS), Knee Injury and Osteoarthritis Outcome Score Physical Function Short Form (KOOS-PS), Knee Outcome Survey Activities of Daily Living Scale (KOS-ADL), Lysholm Knee Scoring Scale, Oxford Knee Score (OKS), Western Ontario and McMaster Universities Osteoarthritis Index (WOMAC), Activity Rating Scale (ARS), and Tegner Activity Score (TAS). Arthritis Care Res 2011; 63(Suppl 11): S208–S228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bůžková P. Linear regression in genetic association studies. PLoS One 2013; 8: e56976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pruim RJ, Welch RP, Sanna S, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics 2010; 26: 2336–2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wessel J. The reliability and validity of pain threshold measurements in osteoarthritis of the knee. Scand J Rheumatol 1995; 24: 238–242. [DOI] [PubMed] [Google Scholar]
- 27.Farrell M, Gibson S, McMeeken J, et al. Pain and hyperalgesia in osteoarthritis of the hands. J Rheumatol 2000; 27: 441–447. [PubMed] [Google Scholar]
- 28.Kosek E, Ordeberg G. Lack of pressure pain modulation by heterotopic noxious conditioning stimulation in patients with painful osteoarthritis before, but not following, surgical pain relief. Pain 2000; 88: 69–78. [DOI] [PubMed] [Google Scholar]
- 29.Arendt-Nielsen L, Nie H, Laursen MB, et al. Sensitization in patients with painful knee osteoarthritis. Pain 2010; 149: 573–581. [DOI] [PubMed] [Google Scholar]
- 30.Boyle AP, Hong EL, Hariharan M, et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res 2012; 22: 1790–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Machiela MJ, Chanock SJ. LDlink: a web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics 2015; 31: 3555–3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Garcia-Etxebarria K, Jauregi-Miguel A, Romero-Garmendia I, et al. Ancestry-based stratified analysis of Immunochip data identifies novel associations with celiac disease. Eur J Hum Genet 2016; 24: 1831–1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Avcin T, Makitie O, Susic M, et al. Early-onset osteoarthritis due to otospondylomegaepiphyseal dysplasia in a family with a novel splicing mutation of the COL11A2 gene. J Rheumatol 2008; 35: 920–926. [PubMed] [Google Scholar]
- 34.Sirko-Osadsa DA, Murray MA, Scott JA, et al. Stickler syndrome without eye involvement is caused by mutations in COL11A2, the gene encoding the alpha2(XI) chain of type XI collagen. J Pediatr 1998; 132: 368–371. [DOI] [PubMed] [Google Scholar]
- 35.Vuoristo MM, Pappas JG, Jansen V, et al. A stop codon mutation in COL11A2 induces exon skipping and leads to non-ocular Stickler syndrome. Am J Med Genet A 2004; 130A: 160–164. [DOI] [PubMed] [Google Scholar]
- 36.Vikkula M, Mariman EC, Lui VC, et al. Autosomal dominant and recessive osteochondrodysplasias associated with the COL11A2 locus. Cell 1995; 80: 431–437. [DOI] [PubMed] [Google Scholar]
- 37.Lapveteläinen T, Hyttinen M, Lindblom J, et al. More knee joint osteoarthritis (OA) in mice after inactivation of one allele of type II procollagen gene but less OA after lifelong voluntary wheel running exercise. Osteoarthritis Cartilage 2001; 9: 152–160. [DOI] [PubMed] [Google Scholar]
- 38.Zeggini E, Panoutsopoulou K, Southam L, et al. Identification of new susceptibility loci for osteoarthritis (arcOGEN): a genome-wide association study. Lancet 2012; 380: 815–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Evangelou E, Kerkhof HJ, Styrkarsdottir U, et al. A meta-analysis of genome-wide association studies identifies novel variants associated with osteoarthritis of the hip. Ann Rheum Dis 2014; 73: 2130–2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Styrkarsdottir U, Thorleifsson G, Helgadottir HT, et al. Severe osteoarthritis of the hand associates with common variants within the ALDH1A2 gene and with rare variants at 1p31. Nat Genet 2014; 46: 498–502. [DOI] [PubMed] [Google Scholar]
- 41.Rodriguez-Fontenla C, Calaza M, Evangelou E, et al. Assessment of osteoarthritis candidate genes in a meta-analysis of nine genome-wide association studies. Arthritis Rheumatol 2014; 66: 940–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Campbell CM, Edwards RR, Fillingim RB. Ethnic differences in responses to multiple experimental pain stimuli. Pain 2005; 113: 20–26. [DOI] [PubMed] [Google Scholar]
- 43.Edwards RR, Doleys DM, Fillingim RB, et al. Ethnic differences in pain tolerance: clinical implications in a chronic pain population. Psychosom Med 2001; 63: 316–323. [DOI] [PubMed] [Google Scholar]
- 44.Edwards RR, Fillingim RB. Ethnic differences in thermal pain responses. Psychosom Med 1999; 61: 346–354. [DOI] [PubMed] [Google Scholar]
- 45.Xu F, Wen T, Lu TJ, et al. Modeling of nociceptor transduction in skin thermal pain sensation. J Biomech Eng 2008; 130: 041013. [DOI] [PubMed] [Google Scholar]
- 46.O'Driscoll SL, Jayson MI. Pain threshold analysis in patients with osteoarthrosis of hip. Br Med J 1974; 3: 714–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.King CD, Sibille KT, Goodin BR, et al. Experimental pain sensitivity differs as a function of clinical pain severity in symptomatic knee osteoarthritis. Osteoarthritis Cartilage 2013; 21: 1243–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]