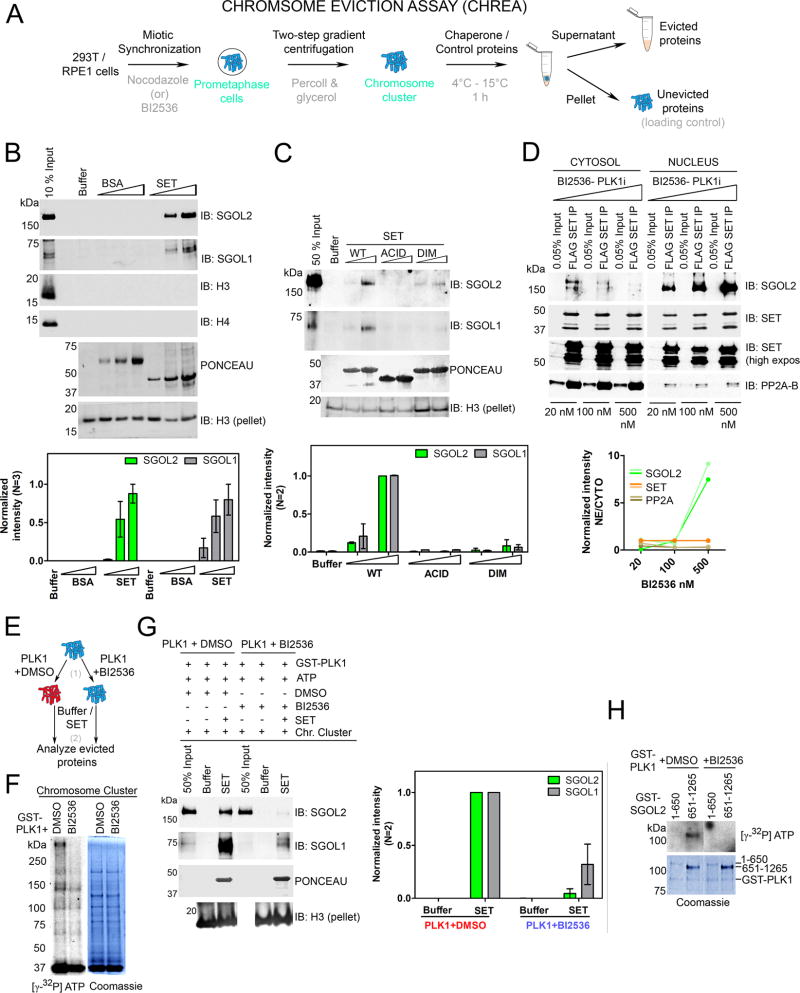
Figure 3. SET is a Shugoshin chaperone and PLK1 augments chaperone activity.
(A) Schematic of CHREA assay to study mitotic protein eviction from chromosomal clusters. (B) (Top) Western blot of evicted proteins reveals that addition of SET, but not BSA, or NAP1 (Figure S3), results in a dose-dependent eviction of SGOL1 and SGOL2 from chromosomes. Additional controls are shown in Figure S3A. Histones H3 and H4 are not evicted. Immunoblot for H3 from the ensuing pellets of the reaction serves as loading control to gauge chromosomal input across samples. (Bottom) H3-pellet and peak normalized intensities from left. (C) (Top) Chaperone assay with SET mutants indicating that both dimerization and acidic tail domains are required for chaperone activity. (Bottom) H3-pellet and peak normalized intensities from left. (D) (Top) NFH-SET RPE1 cells treated with increasing concentrations of BI2536 (20, 100 and 500 nM) display a dose-dependent retention of SET-SGOL2, but not SET-PP2A complexes in the nucleus. (Bottom) Quantification of SET-normalized intensities. Two lines reflect replicates of the experiment. See also Figure S3D for total levels of SGOL2 and SET under these conditions. (E) Schematic of the two-step CHREA in the presence of PLK1. (F) Results of a kinase assay [step 1 of (E)] visualized by autoradiography along with the corresponding Coomassie Blue staining. PLK1 phosphorylates chromosomal clusters used in the chaperone assay. (G) (Left) CHREA performed as in step 2 of (E) indicating that SET evicts SGOL1 and SGOL2 from PLK1 pre-phosphorylated chromosomal clusters, with higher efficiency than those phosphorylated with PLK1+BI2536. (Right) H3-pellet and peak normalized intensities from top. (H) PLK1 kinase assay with purified N-terminal and C-terminal domains of SGOL2. The C-terminal region of SGOL2 is preferentially phosphorylated by PLK1. (See also Figure S3).