Figure 2. In vivo solubilization of DsbB using the SIMPLEx strategy.
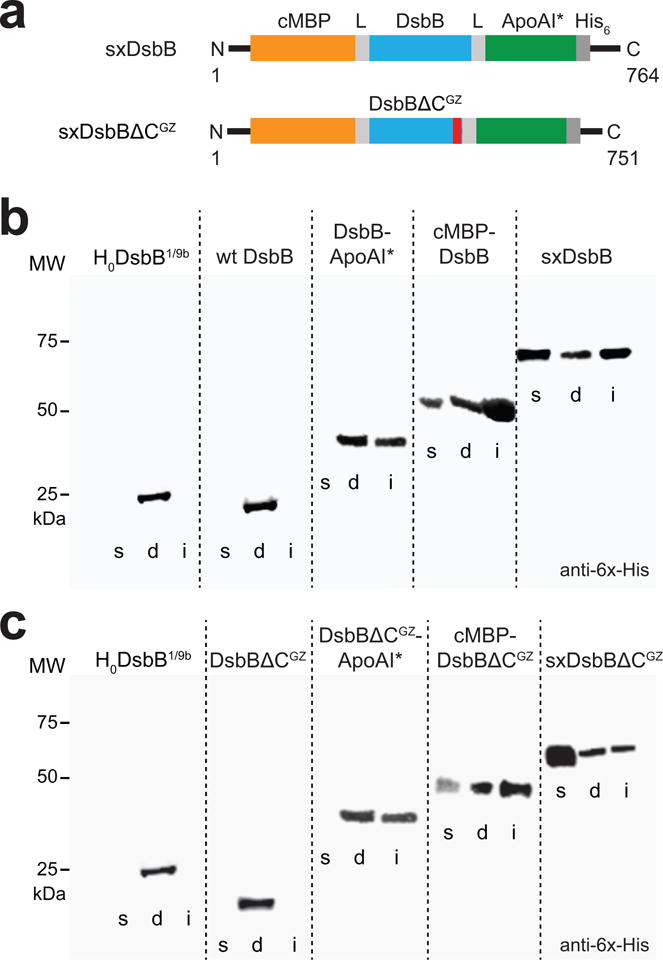
(a) Molecular architecture of SIMPLEx constructs SxDsbB and SxDsbBΔCGZ used in this study. Each construct included N-terminal cMBP as decoy protein (orange), C-terminal ApoAI* as shield domain (green), and intervening flexible linkers (L, grey) that connected cMBP and ApoAI* to the DsbB domain. The DsbB domains tested were wt DsbB (top) and DsbBΔCGZ (bottom), the latter of which is a C-terminally truncated DsbB variant harboring an engineered glycine zipper (GZ, red). (b) Western blot analysis of soluble (s), detergent solubilized (d), and insoluble (i) fractions prepared from E. coli strain BL21(DE3) carrying pET21-based plasmids encoding one of the following DsbB constructs: topologically inverted DsbB (H0DsbB1/9b), wt DsbB, DsbB-ApoAI*, cMBP-DsbB, and cMBP-DsbB-ApoAI* (SxDsbB) as indicated. Blot was probed with anti-6x-His antibody. Molecular weight (MW) markers are shown on the left. (c) Western blot analysis identical to panel (b) except with DsbBΔCGZ in place of wt DsbB in all constructs as indicated. See Supplementary Figure 9 for uncropped versions of the blot images.
