Figure 5. Proper folding of complex substrate proteins by solubilized DsbB variants.
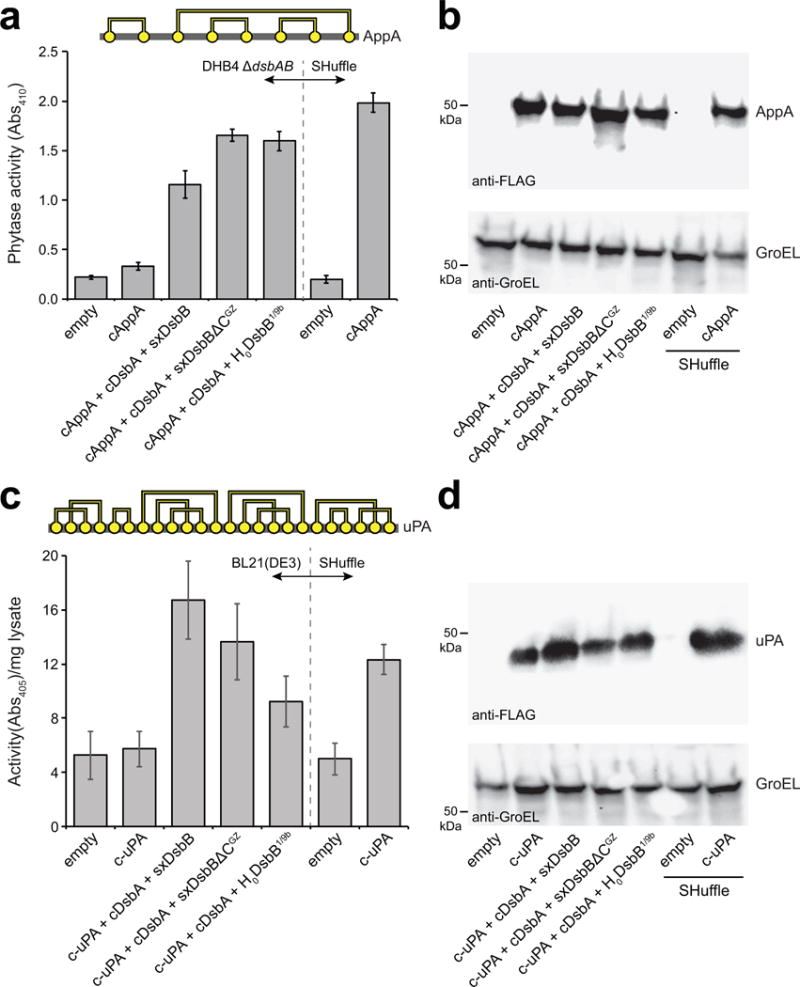
(a) Disulfide bond connectivity for E. coli phytase, AppA (4 disulfide bonds depicted by yellow circles connected by yellow lines). Phytase activity (measured as absorbance at 410 nm) in cytoplasmic fractions derived from: E. coli strain DHB4(DE3) ΔdsbAB carrying no plasmids (empty) or pHIS-cAppA (cAppA) along with pET28-SxDsbB::GST-cDsbA, pET28-SxDsbBΔCGZ::GST-cDsbA, or pET28-H0DsbB1/9b::GST-cDsbA as indicated; and SHuffle cells carrying no plasmid (empty) or pHIS-cAppA (cAppA). (b) Western blot analysis of same cytoplasmic fractions assayed in (a). (c) Disulfide bond connectivity for murine urokinase, uPA (12 disulfide bonds depicted by yellow circles connected by yellow lines). Urokinase activity (measured as absorbance at 405 nm) in cytoplasmic fractions derived from E. coli strain BL21(DE3) carrying no plasmids (empty) or pET24b-urokinase along with pET28-SxDsbB::GST-cDsbA, pET28-SxDsbBΔCGZ::GST-cDsbA, or pET28-H0DsbB1/9b::GST-cDsbA as indicated; and SHuffle cells carrying no plasmid (empty) or pET24b-urokinase. (d) Western blot analysis of same cytoplasmic fractions assayed in (c). Blots were probed with anti-FLAG antibody to detect cAppA and c-uPA (top panels) and anti-GroEL antibody to detect GroEL (bottom panel), which served as a cytoplasmic fractionation marker and loading control. Molecular weight (MW) markers are shown on the left. Activity data is the mean of biological triplicates and the error bars represent the standard error of the mean (SEM). See Supplementary Figure 11 for uncropped versions of the blot images.
