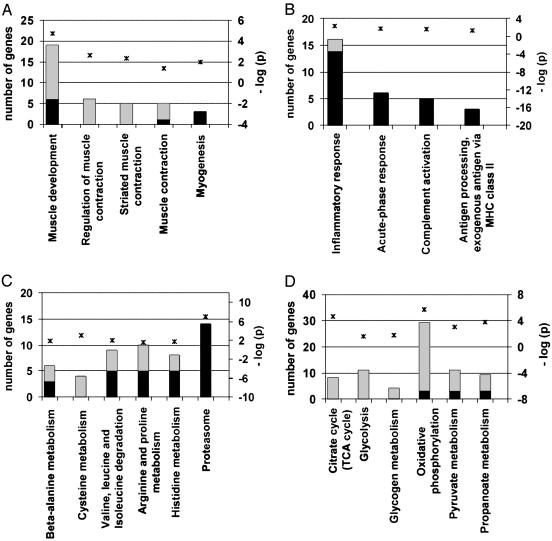
Fig. 1.
Several differentially expressed genes for hind limb burn belong to one of four functional categories, as identified by using Gene Ontology and KEGG metabolic pathways at P ≤ 0.05. Black bars indicate number of up-regulated genes; gray bars correspond to down-regulated genes in the hind limb burn model versus control mice (left y axis). The negative log10 of P values represented by asterisks are indicated in the right y axis.