Figure 3.
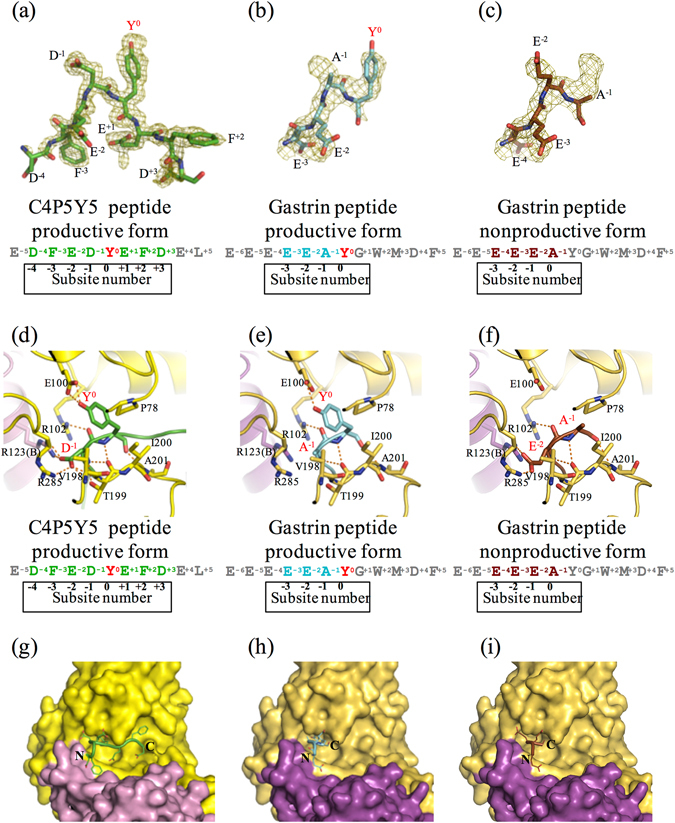
Three substrate peptides in the active site of human TPST1. (a–c) The electron density maps showing the image of C4P5Y5 and the gastrin peptide (simulated annealed omit Fo–Fc maps contoured at 2.5 σ and 1.7 σ, respectively). The Fo–Fc maps are drawn in olive. (a) Fo–Fc map of C4P5Y5. (b) Fo–Fc map of the gastrin peptide productive form. (c) Fo–Fc map of the gastrin peptide nonproductive form. (d–f) Interaction of Y0 and the residue at the subsite -1 in substrate peptides with human TPST1. Hydrogen bonds are depicted as orange dotted lines. The main chain and side chain of Y0 and the residue at the subsite -1 in the substrate peptide are shown. Only the main chain of the other residues in the substrate peptide is shown. Each amino acid sequence is respectively shown below. (d) Human TPST1-PAP-C4P5Y5. (e) The human TPST1-PAP-gastrin peptide productive form. (f) The human TPST1-PAP-gastrin peptide nonproductive form. (g–i) Close-up views of the binding site of C4P5Y5 and the gastrin peptide by surface representation. (g) Human TPST1-PAP-C4P5Y5. (h) The human TPST1-PAP-gastrin peptide productive form. (i) The human TPST1-PAP-gastrin peptide nonproductive form.
