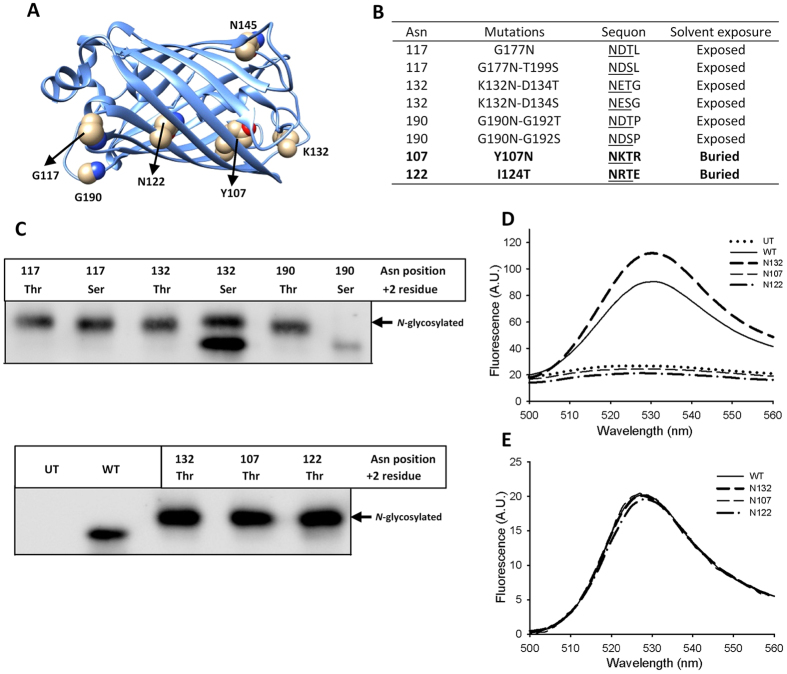
Figure 2.
Buried sequons can be efficiently glycosylated. Yellow florescent protein (YFP) was fused with an N-terminal signal peptide, a C-terminal 3xHA epitope and a KDEL ER retention/retrieval signal. (A) Original residues in YFP (PDB code 1MYW) where Asn in sequons were located. Asn 145 shown in the image was previously used in the N-glycosylation sensor Glyc-ER-GFP27. (B) YFP N-glycosylation variants used in this study. Residues are numbered as in the mature protein after signal peptide cleavage. (C) anti-HA western blot analysis of COS-7 cells expressing variants YFPs. (UT: untransfected cells, WT: wild type non-glycosylatable YFP). Original gels are shown in Supplementary Fig. S5. (D) Emission spectra of transfected COS-7 cells resuspended in lysis buffer. Fluorescence of cells expressing YFP with glycosylation at buried sites sites (variants N107 or N122) is indistinguishable from the fluorescence of untransfected cells. (E) Emission spectra of proteins purified from E. coli (without N-glycosylation).