Abstract
Ligand-dependent corepressor (LCoR) is a transcriptional repressor of ligand-activated estrogen receptors (ERs) and other transcription factors that acts both by recruiting histone deacetylases and C-terminal binding proteins. Here, we first studied LCOR gene expression in breast cancer cell lines and tissues. We detected two mRNAs variants, LCoR and LCoR2 (which encodes a truncated LCoR protein). Their expression was highly correlated and localized in discrete nuclear foci. LCoR and LCoR2 strongly repressed transcription, inhibited estrogen-induced target gene expression and decreased breast cancer cell proliferation. By mutagenesis analysis, we showed that the helix-turn-helix domain of LCoR is required for these effects. Using in vitro interaction, coimmunoprecipitation, proximity ligation assay and confocal microscopy experiments, we found that receptor-interacting protein of 140 kDa (RIP140) is a LCoR and LCoR2 partner and that this interaction requires the HTH domain of LCoR and RIP140 N- and C-terminal regions. By increasing or silencing LCoR and RIP140 expression in human breast cancer cells, we then showed that RIP140 is necessary for LCoR inhibition of gene expression and cell proliferation. Moreover, LCoR and RIP140 mRNA levels were strongly correlated in breast cancer cell lines and biopsies. In addition, RIP140 positively regulated LCoR expression in human breast cancer cells and in transgenic mouse models. Finally, their expression correlated with overall survival of patients with breast cancer. Taken together, our results provide new insights into the mechanism of action of LCoR and RIP140 and highlight their strong interplay for the control of gene expression and cell proliferation in breast cancer cells.
Introduction
Breast cancer is the most common female cancer worldwide.1 Estrogen receptors (ERs) control tumor growth and cell proliferation in about 70% of all breast cancers. ERα and ERβ are members of a large superfamily of transcription factors2 and their action is mediated by transcription co-regulators that act as activators or repressors. Among the repressors, ligand-dependent corepressor (LCoR) shows strong repressive activities upon recruitment by agonist-activated receptors.3, 4
The LCOR gene encodes LCoR, a nuclear protein isolated thanks to its interaction with agonist-activated ERα4 that exerts a strong repressive transcriptional activity partly mediated by recruitment of histone deacetylases, notably histone deacetylase 6,6 and C-terminal binding proteins (CtBPs).5 LCoR corepressor functions are not specific to members of the nuclear receptor family because it also interacts with other transcription factors, such as Kruppel-like factor 67 and KRAB-associated protein 1.8 Moreover, LCoR is a component of a CoREST-CtBP complex that contains the histone H3 demethylase LSD1.9 LCoR also contributes to cell type-dependent and Kruppel-like factor 6-mediated transcriptional repression of the CDKN1A gene that encodes the cell cycle regulator p21WAF1/CIP1.7 Finally, although only few studies on LCoR physiological roles have been published, Asim et al.10 reported that this transcription factor inhibits prostate cancer growth in a xenograft mouse model. Moreover, a recent study demonstrated that LCoR reduces liver triglyceride levels and steatosis in obese mice via repression of thyroid hormone receptor β1 activity on lipogenic gene expression.11
Interestingly, receptor-interacting protein of 140 kDa (RIP140, also called nuclear receptor-interacting protein 1 (NRIP1)) also interacts with agonist-liganded ERα,12 as well as with various nuclear receptors and other transcription regulators.13 RIP140 has an important role in colon cancer.14 It is also required for mammary gland development15 and negatively regulates breast cancer cell proliferation and tumor progression.16, 17 RIP140 shares many similarities with LCoR because it also acts as a transcriptional corepressor that can recruit histone deacetylases and CtBPs.18, 19
In this study, we first analyzed the expression and activity of LCoR and of the LCoR2 variant in breast cancer cells. We then demonstrated the role of the helix-turn-helix (HTH) domain located in LCoR C-terminus in LCoR inhibition of gene expression and breast cancer cell proliferation. We also showed RIP140 role in LCoR-dependent repression of cell proliferation, demonstrated the physical interaction between LCoR and RIP140 and identified the involved domains. Moreover, we unraveled the very strong positive effect of RIP140 on LCoR expression both in mouse mammary tissue and in human breast cancer cell lines and found that their expression was correlated in breast cancer biopsies. Finally, we demonstrated that LCOR and RIP140 gene expression levels have a good prognostic value and are significantly correlated with survival in patients with breast cancer.
Results
LCOR gene expression in breast cancer cells
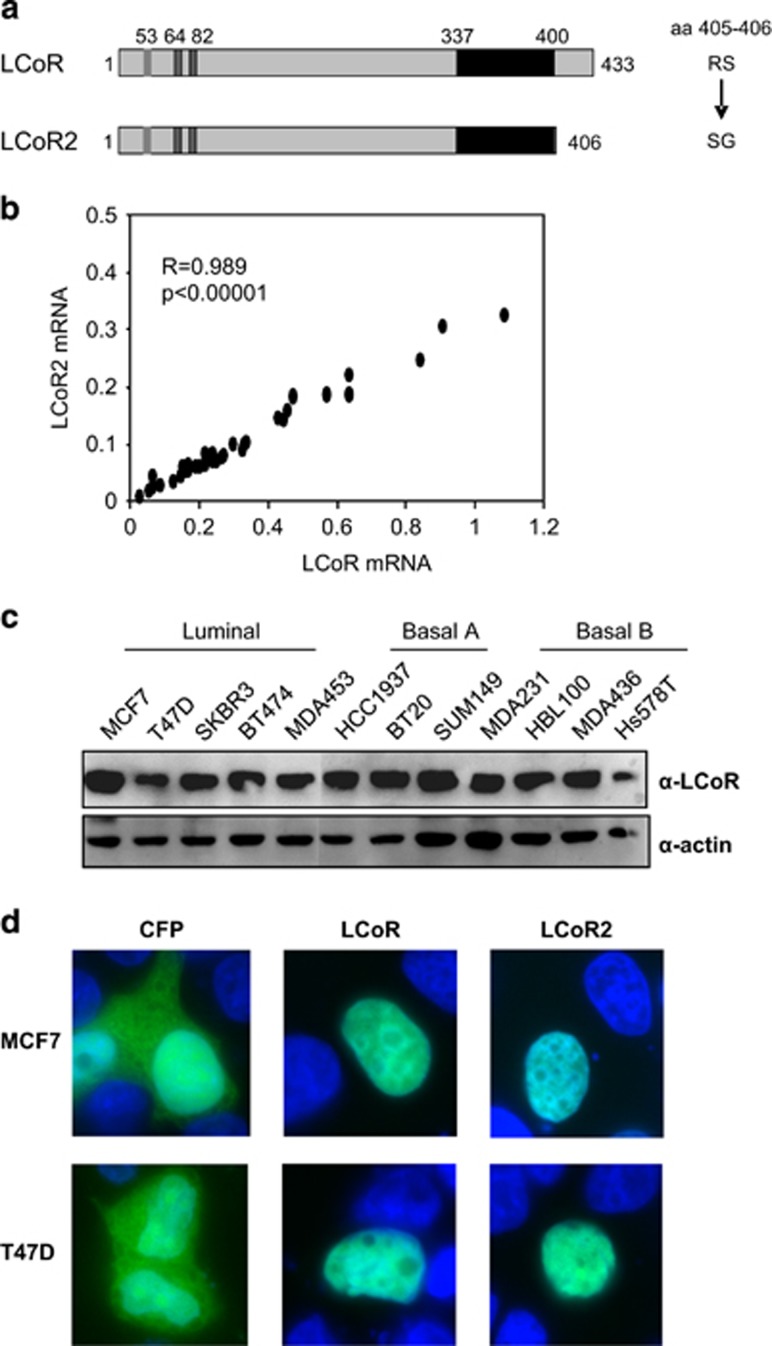
A previous study reported that LCOR is widely expressed in fetal and adult human tissues.4 To better characterize its expression in human mammary epithelial cancer cells, we measured the mRNA levels of two LCoR isoforms in 35 breast cancer cell lines. LCoR2 is a variant initially described by Calderon et al.7 that encodes a protein of 406 residues with a shorter C-terminus (Figure 1a). We found that LCoR and LCoR2 mRNA levels were significantly correlated (r=0.989, P<0.00001; Figure 1b). Western blot analysis indicated that LCoR was expressed in all the tested cell lines, with no significant difference among breast cancer molecular subtypes, although the expression level was slightly lower in T47D and Hs578T cells (Figure 1c). To determine LCoR and LCoR2 subcellular localization, we generated cyan fluorescent protein (CFP)-tagged proteins. Microscopy analysis after transient transfection in MCF7 and T47D cells revealed that the two proteins were expressed in the nucleus (Figure 1d), as previously described for LCoR.5
Figure 1.
LCoR and LCoR2 expression in breast cancer cells. (a) Schematic representation of LCoR and LCoR2 showing the LSKLL motif (aa 53), the CtBP binding site 1 (aa 64) and 2 (aa 82) and the HTH domain (aa 337–400), as well as the two amino acids at position 405 and 406 that differ in LCoR (RS) and LCoR2 (SG). (b) Correlation between LCoR and LCoR2 mRNA expression in 35 breast tumor cell lines. Statistical significance was assessed using the Pearson’s correlation analysis. (c) Whole-cell extracts from the indicated luminal and basal breast tumor cell lines were used for western blot analysis. Membranes were incubated with an anti-LCoR (upper panel) and an anti-actin antibody (lower panel). (d) MCF7 and T47D cells were transfected with CFP, CFP-LCoR or CFP-LCoR2 expressing plasmids. Two days after transfection, cells are fixed with 4% paraformaldehyde and then observed using an AxioImager Z2 microscope.
LCoR and LCoR2 inhibit gene expression
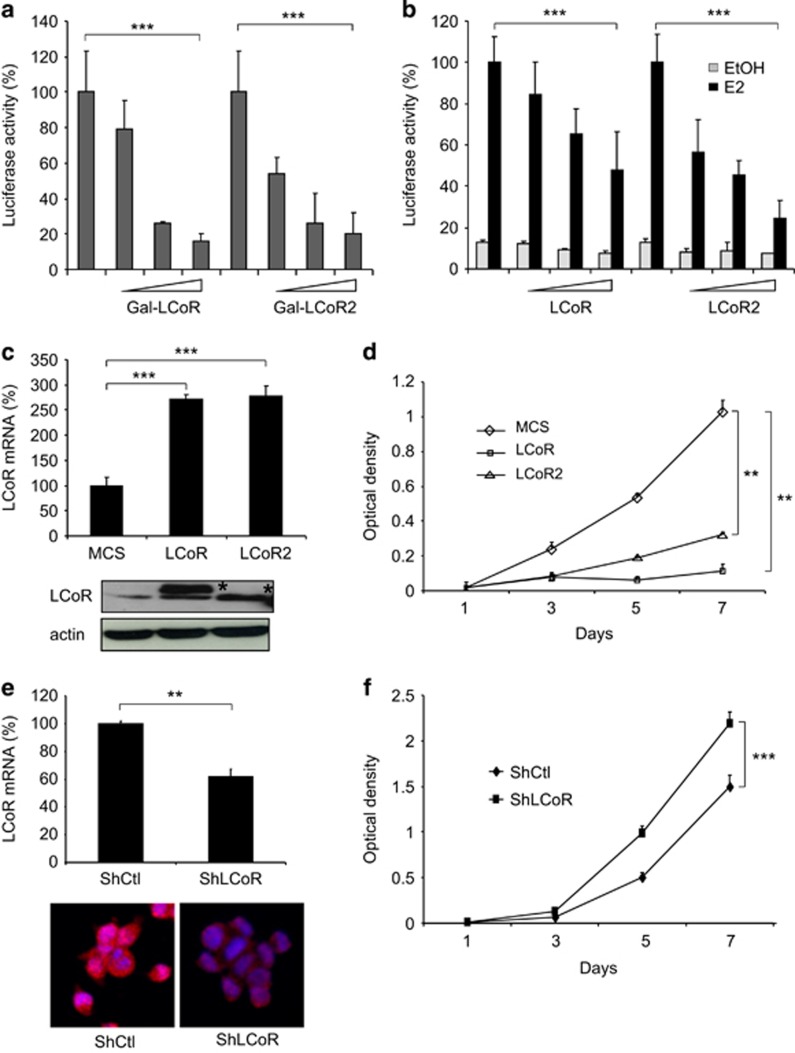
To better characterize LCoR and LCoR2 biological properties, we first investigated their transcriptional activity in the T47D luminal breast cancer cell line. GAL4DBD-LCoR and LCoR2 fusion proteins displayed a strong intrinsic repressive activity (Figure 2a), as already reported for LCoR.4 The two proteins repressed, in a dose-dependent manner, the estrogen (E2)-dependent transcriptional activity of an estrogen response element (ERE)-driven reporter gene and, in these conditions, LCoR2 appeared to be a stronger repressor (Figure 2b).
Figure 2.
LCoR and LCoR2 activity in breast cancer cells. (a) T47D cells were transiently transfected with the pRL-CMV-renilla and L8G5 reporter plasmids, LexAVP16 expression plasmid and increasing doses of Gal4DBD-LCoR and -LCoR2 expression plasmids. Luciferase values were normalized to the renilla luciferase control and expressed as the percentage of Gal4DBD activity. ***P<0.001. (b) T47D cells were transiently transfected with pEBL+ (encoding ERE-driven luciferase), pRL-CMV-renilla as internal control, and increasing doses of p3XFlag-LCoR and p3XFlag-LCoR2 expression vectors. Cells were treated with vehicle (EtOH) or 10−8 m estradiol (E2) for 16h. Luciferase values were normalized as in a and expressed as the percentage of the activity in the absence of transfected LCoR. ***P<0.001. (c) T47D cells were stably transfected with LCoR or LCoR2-expressing plasmids, or vector alone (MCS). LCoR and LCoR2 mRNA and protein expression levels were checked by real-time PCR (upper panel) and by western blotting with an anti-Flag and an anti-actin antibody (lower panels), respectively. (d) Cell proliferation was measured over four days. Absorbance of formazan crystals was read on a spectrophotometer and results were normalized to the cell density at day 1. (e) T47D cells were stably transfected with short hairpin-expressing plasmids against LCoR (ShLCoR) or a control sequence (ShCtl). LCoR mRNA and protein expression levels were then checked by real-time PCR (upper panel) and by immunofluorescence using anti-LCoR antibody and Hoechst staining (lower panels), respectively. (f) Cell proliferation was measured over 4 days using cells from e. Results were normalized as in d. Statistical analyses were performed using the Mann–Whitney test. **P<0.01; ***P<0.001.
LCoR and LCoR2 inhibit cell proliferation
Then, to investigate LCoR and LCoR2 effect on cell proliferation, we stably transfected T47D cells with plasmids that encode LCoR or LCoR2. In transfected cells, LCoR and LCoR2 mRNA (Figure 2c, upper panel) and protein (lower panel) expression levels were increased compared with cells transfected with empty vector multiple cloning site (MCS). Moreover, the E2-dependent induction of the estrogen-regulated gene GREB1 was significantly reduced in both LCoR and LCoR2-expressing cells, compared with MCS cells (Supplementary Figure S1A), further indicating that these T47D cell lines express functional proteins. 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay revealed that the T47D cells that stably express LCoR or LCoR2 displayed a severe proliferation defect, compared with control cells (MCS) (Figure 2d). In parallel, T47D cells were stably transfected with vectors to express an LCoR-targeting shRNA (ShLCoR) or a control shRNA (ShCtl). Endogenous LCoR mRNA and protein expression was reduced in ShLCoR-expressing cells compared with ShCtl cells (Figure 2e). Endogenous GREB1 expression reduction upon E2 stimulation in ShLCoR-expressing cells further confirmed the functional downregulation of endogenous LCoR (Supplementary Figure S1B). By using three different techniques, including MTT assay (Figure 2f), impedance measurement (Supplementary Figure S2A) and cell counting (Supplementary Figure S2B), we showed that cell proliferation was higher in ShLCoR-expressing cells compared with ShCtl cells, strengthening the results obtained by ectopic expression of LCoR. Similar results were observed also by knocking-down LCoR expression in MCF7 cells (Supplementary Figures S3A and B).
Role of the HTH domain in LCoR activities
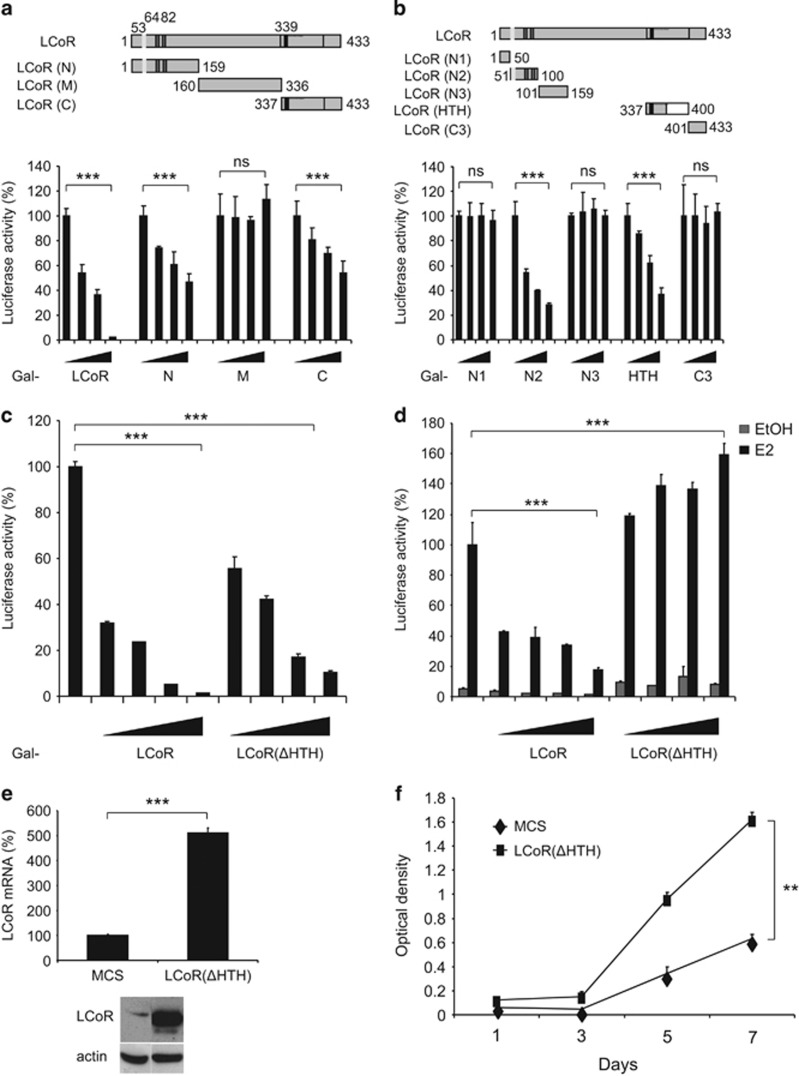
To identify LCoR repressive domains, we generated LCoR mutants that were expressed as GAL4DBD fusion proteins (Figure 3a, upper panel). Both the N-terminal domain N (aa 1–159) and the C-terminal domain C (aa 337–433) displayed a strong intrinsic repressive activity, whereas the central domain M (aa 160–336) presented no activity (Figure 3a, lower panel). To further define the inhibitory regions, we created shorter domains within the N and C-terminal regions (Figure 3b, upper panel). The N2 (aa 51–100) and the HTH domains (aa 337–400) showed a strong repressive activity. Conversely, domains N1 (aa 1–50), N3 (aa 101–159) and C3 (aa 401–433) had virtually no inhibitory activity. We concluded that LCoR possesses two independent repressive domains: N2 that harbors the CtBP-interacting motifs and the HTH region.
Figure 3.
The HTH domain has a major role in LCoR activity. (a, b) Upper panels. Schematic representation of LCoR fragments. Lower panels. COS7 cells were transiently transfected with pRL-CMV-renilla and L8G5 reporter plasmids, LexAVP16 and increasing doses of plasmids expressing the various LCoR variants fused to Gal4DBD. Luciferase values were expressed as described in Figure 2a. (c) T47D cells were transiently transfected with pRL-CMV-renilla and L8G5 reporter plasmids, LexAVP16 and increasing doses of Gal4DBD-LCoR or -LCoR(ΔHTH) expression plasmids. Luciferase values were normalized as described in Figure 2a. (d) T47D cells were transiently transfected with pEBL+ (encoding ERE-driven luciferase), pRL-CMV-renilla as internal control and increasing doses of p3XFlag-LCoR or p3XFlag-LCoR(ΔHTH) expression vectors. Cell treatment and data normalization were as in Figure 2b. (e) T47D cells were stably transfected with LCoR(ΔHTH). LCoR(ΔHTH) mRNA and protein expression were checked by real-time PCR (upper panel) and western blotting with an anti-Flag and an anti-actin antibody (lower panels), respectively. (f) Cell proliferation was measured as described in Figure 2d. Statistical analyses were performed using the Mann–Whitney test. NS, not significant; **P<0.01; ***P<0.001.
To better understand the role of the HTH domain, we generated a construct to express an LCoR mutant without this domain (LCoR(ΔHTH)) fused to GAL4DBD. Gal-LCoR(ΔHTH) exhibited a significantly weaker repressive activity compared with wild-type Gal-LCoR (Figure 3c). Moreover, LCoR(ΔHTH) did not inhibit ERE-dependent transcriptional activity, but rather stimulated luciferase activity (Figure 3d). We next showed that in T47D cells that stably express LCoR(ΔHTH) (Figure 3e), cell proliferation was significantly increased compared with MCS cells (Figure 3f). These results show that the HTH domain is required for cell proliferation inhibition by LCoR.
RIP140 is necessary for LCoR repressive activity
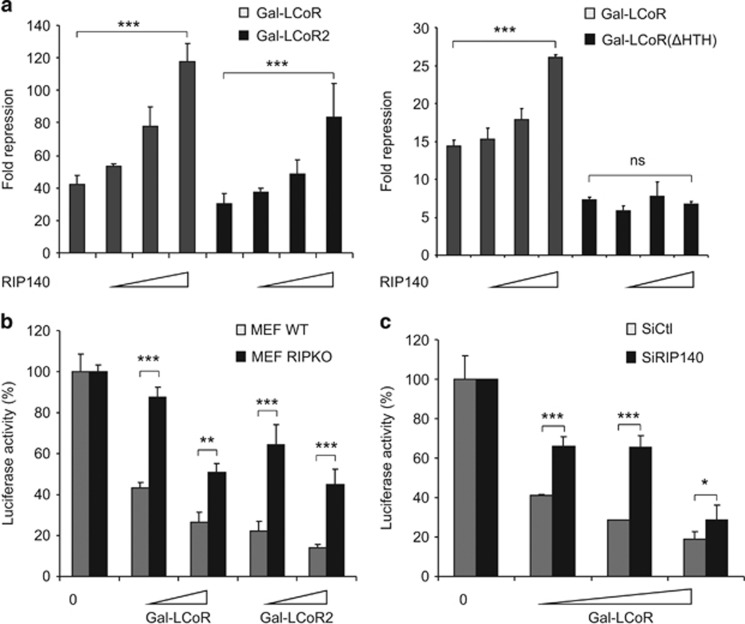
Similarly to LCoR, RIP140 was isolated thanks to its interaction with ERα, interacts with CtBP and histone deacetylase proteins and is involved in the regulation of breast cancer cell proliferation.20 We thus investigated the possible cross-talk between LCoR and RIP140 in breast cancer cells. In transient transfection experiments, RIP140 drastically increased the repressive activity of both Gal-LCoR and Gal-LCoR2 (Figure 4a, left panel). Conversely, it did not have any effect on Gal-LCoR(ΔHTH) activity (Figure 4a, right panel).
Figure 4.
RIP140 is necessary for full LCoR-mediated transcriptional repression. (a) COS7 cells were transiently transfected with pRL-CMV-renilla and L8G5 reporter plasmids, LexAVP16, Gal4DBD-LCoR or -LCoR2 (left panel) or Gal4DBD-LCoR and -LCoR(ΔHTH) (right panel) expression plasmids and increasing concentrations of c-Myc-RIP140 vector. Luciferase values were normalized as in Figure 2a and expressed as the ratio between Gal4DBD alone and Gal4DBD-LCoR. (b) Wild-type (WT) and RIP140 knock-out (RIPKO) MEFs were transiently transfected with pRL-CMV-renilla and L8G5 reporter plasmids, LexAVP16 and increasing doses of Gal4DBD-LCoR and -LCoR2 expression plasmids as indicated. Luciferase values, normalized to the renilla luciferase control, were expressed as the percentage of the activity obtained in cells transfected with Gal4DBD expression plasmid. (c) MCF7 cells were transiently transfected with pRL-CMV-renilla, L8G5 reporter plasmids, LexAVP16, increasing doses of Gal4DBD-LCoR and either siCtl or siRIP140 siRNAs, as indicated. Luciferase values were expressed as in b. Statistical analyses were performed using the Mann–Whitney test. NS, not significant; *P<0.05; **P<0.01; ***P<0.001.
In mouse embryo fibroblasts (MEFs), Gal4-LCoR and -LCoR2 displayed a strong intrinsic repressive activity (Figure 4b), as expected. Conversely, in RIP140 knock-out (RIPKO) MEFs, the repressive activity of Gal-LCoR and Gal-LCoR2 was significantly reduced, indicating that RIP140 is necessary for their full repressive potential. This was confirmed in MCF7 cells transfected with increasing amounts of Gal4-LCoR and with a small interfering RNA (siRNA) targeting RIP140 (Figure 4c). The strong transcriptional repressive activity of Gal-LCoR was significantly affected upon RIP140 downregulation (compared with control siRNA), further reinforcing the results obtained in MEFs. Altogether, these data clearly demonstrate that RIP140 strongly influences LCoR-mediated gene expression repression.
RIP140 regulates LCoR-dependent cell proliferation
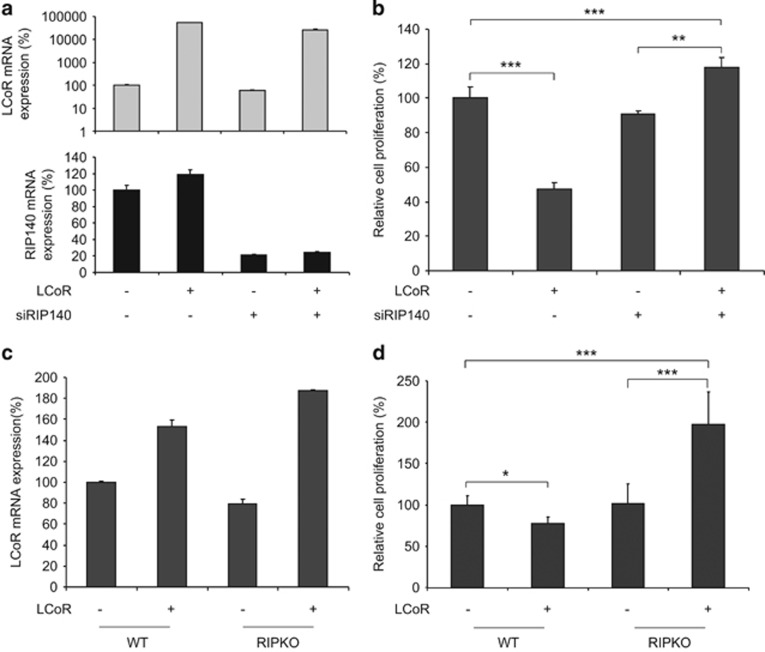
As RIP140 controls LCoR transcriptional activity, we asked whether it could also modulate LCoR effect on cell proliferation. We first transiently transfected T47D cells with an LCoR-expressing plasmid and a control (siCtl) or a RIP140-specific (siRIP140) siRNA. Cells transfected with the LCoR-expressing vector showed a significant increase in LCoR mRNA expression, whereas those transfected with the siRIP140 siRNA showed an 80% decrease in RIP140 mRNA expression (Figure 5a). As expected, LCoR overexpression inhibited cell proliferation (Figure 5b). Conversely, in cells in which LCoR was overexpressed and RIP140 mRNA was concomitantly downregulated, cell proliferation was strongly increased compared with cells that express only LCoR. In parallel, the mRNA levels of the proliferation marker Ki67 and the cell cycle regulator p21 were quantified and they perfectly matched the data obtained on cell proliferation (Supplementary Figures S4A and B, respectively). Moreover, concomitant LCoR overexpression and RIP140 downregulation in HCC1937 basal breast cancer cells also led to an increase in cell proliferation (Supplementary Figure S5A). As p21 was deregulated upon LCoR ectopic expression, we wondered whether its expression would affect LCoR-dependent cell proliferation. We thus used MEFs expressing wild-type or knock-out for P21 gene20 and transfected them with an LCoR-expressing vector. As shown in Supplementary Figure S5B, LCoR expression resulted in a decreased proliferation of WT MEFs, whereas p21KO MEFs expressing LCoR displayed an increased proliferation. This experiment depicted a direct link between p21 cell status and LCoR-mediated cell proliferation.
Figure 5.
RIP140 modulates LCoR-mediated cell proliferation. (a) T47D cells were co-transfected with a Flag or a Flag-LCoR-expressing vector together with siCtl or siRIP140 siRNAs, as indicated. RIP140 and LCoR mRNA levels were quantified by real-time PCR. Results were corrected to 28S mRNA expression and normalized to the values from cells transfected with the control plasmid and siRNA. (b) Cell proliferation was measured over 10 days. Absorbance of formazan crystals was read on a spectrophotometer and cell proliferation at day 10 was expressed relative to control (empty expression vector and siCtl). (c) Wild-type (WT) and RIPKO MEFs were stably transfected with a GFP or a GFP-LCoR-expressing plasmid. LCoR mRNA levels were quantified as in a. (d) Cell proliferation was assayed as described in (b). *P<0.05; **P<0.005; ***P<0.001.
To strengthen these results, we then stably transfected wild-type and RIPKO MEFs with a green fluorescent protein (GFP)- or a GFP-LCoR-expressing vector. MEFs transfected with the GFP-LCoR plasmid showed stronger LCoR mRNA expression than those transfected with GFP (Figure 5c). As before, LCoR overexpression in wild-type MEFs reduced cell proliferation compared with MEFs that express only GFP (Figure 5d). Remarkably, LCoR overexpression in RIPKO MEFs significantly promoted cell proliferation, in line with the results obtained in human breast cancer cell lines. Altogether, these two series of data indicate that RIP140 is necessary for the full repressive effect of LCoR on cell proliferation.
RIP140 interacts with LCoR
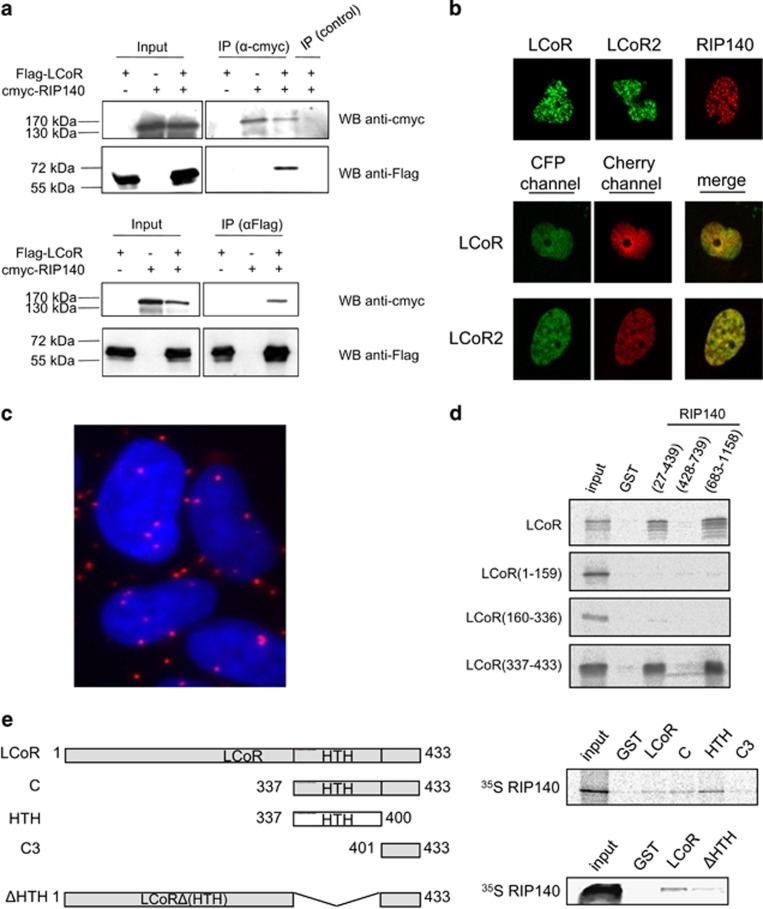
We next asked whether RIP140 and LCoR could be part of the same protein complex. Immunoprecipitation of tagged RIP140 from COS7 cells that ectopically express c-Myc-RIP140 and/or Flag-LCoR could pull-down also Flag-LCoR, when the two proteins were co-expressed (Figure 6a, upper panels). The interaction was specific because an unrelated antibody failed to immunoprecipitate RIP140 and LCoR. Similarly, the anti-Flag antibody could immunoprecipitate LCoR and also c-Myc-RIP140, when co-expressed (Figure 6a, lower panels), further confirming that the two proteins can interact in a cellular context.
Figure 6.
LCoR interacts with RIP140. (a) LCoR and RIP140 coimmunoprecipitate. COS7 cells were transfected with Flag-LCoR and/or c-Myc-RIP140 expression plasmids, as indicated. Two days after transfection, whole-cell protein lysates were immunoprecipitated with the anti-c-Myc, the anti-Gal4DBD antibody (control) (upper panels) or the anti-Flag antibody (lower panels) and analyzed by western blotting using anti-c-Myc or anti-Flag antibodies. An aliquot of each extract was also immunoblotted to evaluate the expression levels of LCoR and RIP140 (input). (b) COS7 cells were transfected with CFP-LCoR, CFP-LCoR2 and/or Cherry-RIP140 expression plasmids, as indicated. Two days after transfection, cells are analyzed as in Figure 1c. (c) In situ proximity ligation assay between LCoR and RIP140 in MCF7 cells using anti-RIP140 and anti-LCoR antibodies diluted in phosphate-buffered saline–1% bovine serum albumin. (d) GST pull-down assays were carried out using bacterially expressed GST and GST-RIP140 fragments and 35S-labeled LCoR and LCoR deletion mutants. (e) Schematic representation of full-length and LCoR deletion mutants (left). GST pull-down assays were carried out as in d with GST and the indicated GST-LCoR fragments and 35S-labeled RIP140 (right).
Confocal microscopy using COS7 cells transfected with CFP-tagged LCoR or LCoR2 or Cherry-tagged RIP140 indicated that LCoR and RIP140 formed nuclear foci (Figure 6b, upper panels). When LCoR or LCoR2 was co-expressed with RIP140, these proteins did not form nuclear foci, but displayed a diffuse nuclear localization. Moreover, when the CFP and the Cherry channels were merged, the two images could be superimposed, indicating that RIP140 and LCoR/LCoR2 colocalized (Figure 6b, middle and lower panels).
To determine whether endogenous LCoR and RIP140 interacted, we next performed proximity ligation assays in MCF7 breast cancer cells, which confirmed the interaction between the two transcription factors (Figure 6c). Interestingly, we also described an interaction between endogenous LCoR and RIP140 in two other breast cancer cell lines, that is, T47D and HCC19317 (Supplementary Figure S6A). We observed many nuclear and cytoplasmic dots, whereas CFP-LCoR and Cherry-RIP140 were exclusively nuclear. These data prompted us to perform immunofluorescence experiments to investigate the localization of the endogenous proteins. LCoR expression was evenly distributed between cytoplasm and nucleus, whereas RIP140 was mostly nuclear with discrete foci and exhibited only a faint cytoplasmic expression (Supplementary Figure S6B).
To identify the LCoR and RIP140 domains involved in the interaction, we performed GST pull-down experiments with various deletion mutants. We found that the N-terminal (27–439) and C-terminal (683–1158) domains of RIP140 interacted with the C-terminal region (337–433) of LCoR (Figure 6d). Moreover, the HTH domain of LCoR pulled-down full-length RIP140, whereas the LCoR(ΔHTH) mutant interacted only weakly with RIP140 (Figure 6e). These results suggest that the HTH domain of LCoR is the major region for interaction with RIP140.
RIP140 regulates LCoR gene expression
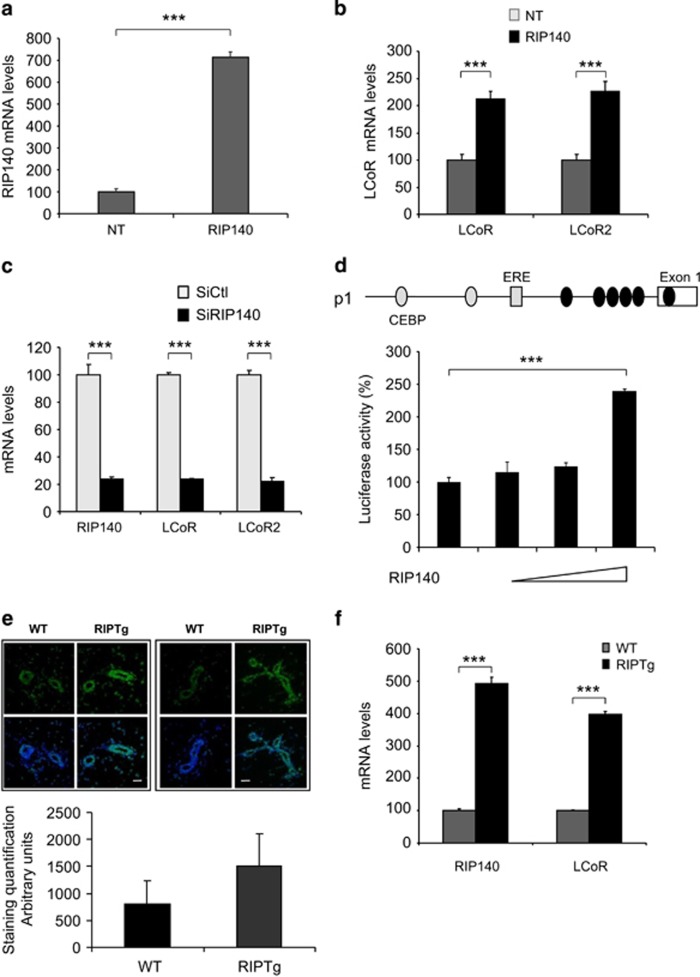
As LCoR and RIP140 can interact and RIP140 is required for LCoR repressive activity, we next asked whether RIP140 influenced LCoR expression. Transient transfection of a RIP140 expression vector in MCF7 cells, resulted in a strong increase of RIP140 mRNA levels (Figure 7a) and in a two-fold upregulation of both LCoR and LCoR2 mRNA expression (Figure 7b), compared with non-transfected (NT) cells. This suggests the existence of a positive regulation of LCoR expression by RIP140. Conversely, upon siRNA-mediated RIP140 downregulation, the expression of both LCoR and LCoR2 was also severely reduced (Figure 7c). To analyze RIP140 effect at the transcriptional level, we transfected increasing doses of the RIP140 expression vector and measured LCoR promoter activity. RIP140 could stimulate the luciferase activity driven by the LCoR promoter in a dose-dependent manner (Figure 7d).
Figure 7.
LCoR expression is regulated by RIP140. (a, b) MCF7 cells were transfected or not (NT) with the c-Myc-RIP140 expressing plasmid. Two days after transfection, total RIP140 (a), LCoR and LCoR2 (b) mRNA levels were quantified by real-time PCR. Results were corrected to 28S mRNA expression and normalized to those of NT MCF7 cells. (c) Two days after transfection of Control (siCtl) and RIP140 siRNA, RIP140 and LCoR mRNAs were measured by real-time PCR. Results were corrected to 28S mRNA expression and normalized to those of control siRNA transfected cells. (d) Schematic representation of the LCoR promoter (p1) (upper panel). MCF7 cells were transiently transfected with 50 ng of pRL-CMV-renilla, 50 ng of p1-luc and increasing concentrations of c-Myc-RIP140 expression plasmids. Luciferase values were expressed as described in Figure 2a. (e) LCoR labeling (green) in paraffin-embedded sections of mouse mammary ducts (left panels) and terminal end buds (right panels) of wild-type (WT) and transgenic mice that overexpress RIP140 (RIPTg). Upper panel: LCoR (green) and nuclear staining (blue). Scale bar, 10 μm. Lower panel: quantification of LCoR staining in WT and RIPTg tissue sections. (f) Expression of RIP140 and LCoR mRNA was measured by real-time PCR in mammary glands from WT and RIPTg mice. Values were normalized to RS9 mRNA and expressed relative to the values in WT mice.
To strengthen these results, we then analyzed LCoR expression in mouse mammary glands from wild-type animals and transgenic mice that overexpress RIP140 (RIPTg). LCoR signal intensity in epithelial cells of both ducts and terminal end buds was significantly higher in RIPTg than in wild-type samples (Figure 7e), supporting the hypothesis that RIP140 regulates LCoR expression. These results were confirmed by quantification of LCoR mRNA expression in wild-type and RIPTg mammary glands (Figure 7f).
This series of data support a robust RIP140 effect on LCOR gene expression.
LCoR and RIP140 expression in human breast cancers
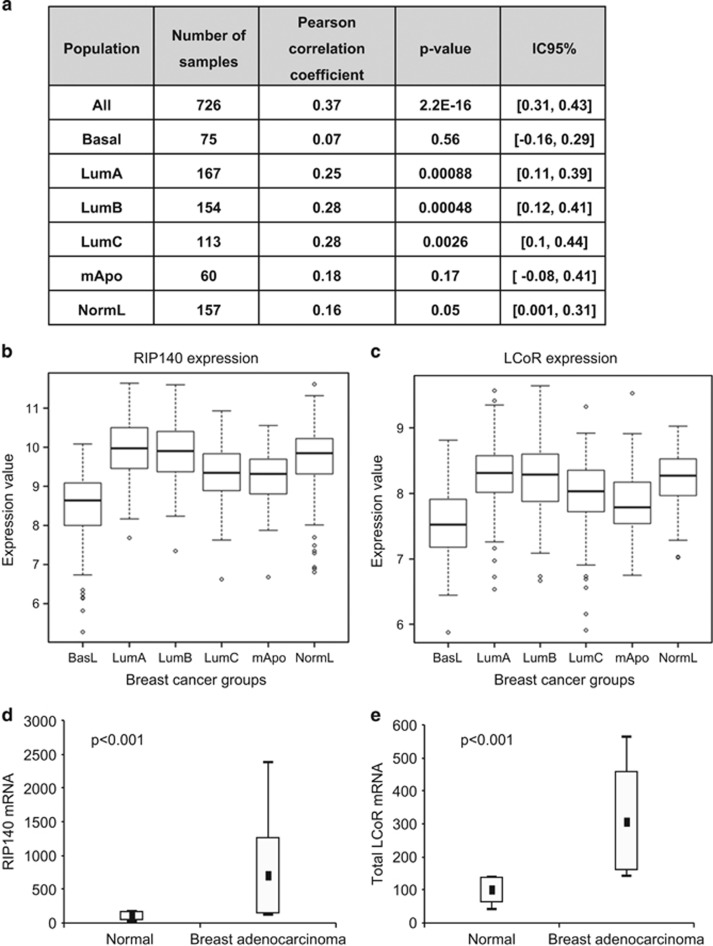
To validate the results obtained in human breast cancer cell lines, we analyzed LCoR and RIP140 expression in human breast cancer biopsies. LCoR and RIP140 mRNA expression were correlated in a cohort of 726 breast cancer tumors (r=0.37; P=2.2e−16) (Figure 8a). Similar results were obtained by reverse transcriptase–quantitative PCR analysis of 35 human breast cancer cell lines (Supplementary Figure S7A) and of an independent series of 135 breast cancer samples (r=0.695, P<0.00001, data not shown). Both RIP140 (Figure 8b) and LCoR (Figure 8c) were strongly expressed in luminal A, luminal B and normal-like tumors. Their expression was markedly lower in basal-like tumors as well as in mApo and luminal C tumors that have the worst prognosis.21 The Breast Cancer Gene-Expression Miner v3.2 correlation module22 confirmed a significant correlation between the expression levels of the two genes in 3288 breast cancer samples (r=0.38; P=0.0001) (Supplementary Figure S7B). Moreover, RIP140 (Figure 8d) and LCoR (Figure 8e) mRNA expression were significantly higher in breast adenocarcinoma samples than in normal tissue. When we took into account the expression of both transcription factors, we found that LCoRlow/RIP140low expression was associated significantly with the most aggressive tumors (grades 2 and 3; Supplementary Figure S7C).
Figure 8.
LCoR and RIP140 are co-expressed in breast cancer. (a) Correlation between RIP140 and LCoR expression in 726 breast tumors and in the subgroups corresponding to the different molecular subtypes.21 The number of samples, Pearson correlation coefficient, P-values and confidence interval are indicated. (b) Box plots showing RIP140 expression in breast cancer samples according to the molecular subtypes. (c) Same as in b for LCoR expression. (d) RIP140 mRNA expression was quantified in a plate containing two identical sets of 43 breast carcinomas and 5 normal tissues. Data are presented as box plots showing the median value, upper and lower quartiles, minimum and maximum non-atypical values. Results were normalized to 28S mRNA expression. (e) Same as in d for LCoR mRNA expression.
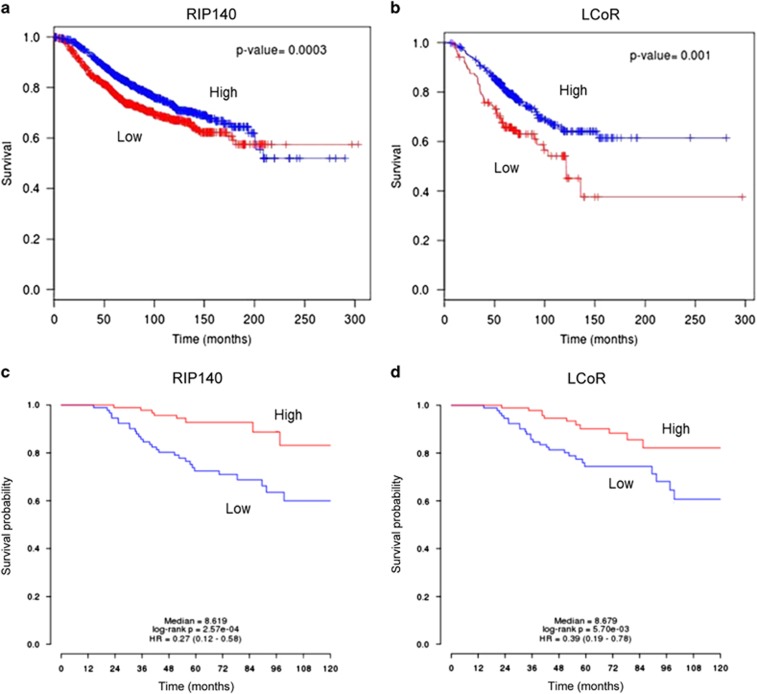
To investigate whether LCoR and RIP140 mRNA levels were associated with patient survival, we used the BreastMark database to reanalyze transcriptomic data sets in order to correlate gene expression levels with clinical outcome. Low LCoR (n=485; P=0.001) and RIP140 (n=2091; P=0.00036) levels were correlated with shorter overall survival in patients with breast cancer (Figures 9a and b). We confirmed these results by using the PRECOG database to analyze the GEO data set GSE24450.23 In this cohort of 183 patients with breast cancer, high LCoR and RIP140 mRNA levels were also significantly associated with better survival (Figures 9c and d). Altogether, these results show that LCoR and RIP140 expression levels have a prognostic value in patients with breast cancer.
Figure 9.
Correlation between RIP140 and LCoR expression and patient survival. The BreastMark algorithm was used to perform on-line Kaplan–Meier analyses of several data sets. (a) The analysis was performed according to RIP140 expression levels (median cut-off) in 2091 breast cancer samples. (b) Same analysis performed according to LCoR expression with a low cut-off in 485 breast cancer samples. The P-values are indicated. Data concerning the correlation between RIP140 (c) and LCoR (d) expression with survival in the cohort of 183 patients with breast cancer (GSE24450)23 were extracted from the PRECOG database using a median cut-off.
Discussion
ER signaling has a major role in the regulation of breast cancer growth and the molecular actors involved in the regulation of this pathway need to be better characterized in order to improve breast tumor phenotyping and the efficacy of ER-targeted therapies, such as tamoxifen or aromatase inhibitors.
In this study, we demonstrate the links between two major transcriptional regulators of ER activity: LCoR and RIP140. Using various approaches (GST pull-down, coimmunoprecipitation, proximity ligation assays and confocal microscopy), we show that LCoR and RIP140 directly interact and are part of a nuclear protein complex. Recent studies have extensively described the complex regulation of RIP140 activity by post-translational modifications (see Yang and Seto24 for a review). It will be of interest to know whether they also modulate its interaction with LCoR for which no modification has been described yet.
Moreover, the RIP140 cistrome has been characterized in MCF7 breast cancer cells25 and it will be interesting to perform ChIP-Seq experiments to determine which genes targeted by RIP140 also recruit LCoR and to identify, among the transcription factors that are regulated by RIP140 or LCoR (including E2Fs,20 NF-κB26 and Kruppel-like factor 6),7 the molecules that may be modulated by the RIP140–LCoR complex.
Our results on the role of the HTH domain of LCoR in its inhibitory activity are supported by previous observations showing that the disruption of this domain markedly attenuates corepression of progesterone receptor and ERα by LCoR.5 Whole-genome sequencing and experimental work highlighted the ubiquity and central role of this domain in gene regulation in both prokaryotes and eukaryotes.27 The solution structure of the HTH domain of human LCoR has been determined (PDB ID: 2cob). Mimetic peptides that can compete with RIP140 for binding to LCoR HTH domain could help understanding the biological relevance of the RIP140-–LCoR interaction.
This study demonstrates LCoR repressive activity on breast cancer cell proliferation and uncovers the major role of the RIP140–LCoR complex in the regulation of this function. Intriguingly, loss of RIP140 expression switches LCoR effect from inhibition of proliferation to its promotion. LCoR negatively regulates the expression of the CDKN1A gene.7 We further showed that along with RIP140, P21 gene expression could switch LCoR activity from a repression to a stimulation of cell proliferation. Interestingly, p21WAF1/CIP1 has both tumor-suppressive and oncogenic properties, depending on the cellular context. For instance, in breast cancer, it can show mitogenic activity (Abbas and Dutta28 and references herein) thus providing a potential mechanistic explanation for the negative effect on cellular proliferation. Most importantly, the cell cycle regulator p21 has been proposed to function as a nodal point for signals from many factors with opposing functions in cancer and as a switch that determines the outcome of these signaling pathways. Our data indicate that p21 expression regulation by LCoR is switched by RIP140 and further work is needed to provide the molecular basis for these regulations and to determine their functional relevance in cell growth regulation by the RIP140-LCoR complex.
Finally, we assessed the physiological relevance of the interaction between RIP140 and LCoR. RIP140 biological importance has been investigated using mice models with loss (RIPKO) or gain (RIPTg) of function. RIPKO mice are viable, but their phenotype indicates that RIP140 has important roles in various tissues and organs, such as ovaries, mammary glands, intestine, muscle, heart and the central nervous system.29, 30, 31, 32, 33, 34 Understanding LCoR effect on RIP140, physiopathological effects will require the generation of transgenic mice models that lack or overexpress LCoR in order to cross them with RIP140 transgenic mice.
Based on our finding about LCoR inhibition of breast cancer cell proliferation, we did not expect to detect higher LCoR mRNA expression in breast cancer cells/tissues compared with normal samples. This result is reminiscent of the work by Poulard et al.30 showing that JMJD6 is highly expressed in breast cancer, although it has anti-proliferative effect in MCF7 cells.
Finally, we found that LCOR and RIP140 gene expression levels are highly correlated in breast cancers and significantly associated with patient survival. Further studies are required to determine whether these two genes are new biological markers that could predict the response to targeted therapies used to treat patients with breast cancer.
Materials and methods
Plasmids
pRL-CMV-renilla and pGL promoter were obtained from Promega (Charbonnieres, France). L8G5-luc and LexAVP16 are kind gifts of Dr Khochbin (INSERM U823, Grenoble, France). The R900 plasmid was described elsewhere.31 ERE-β-globin-luciferase (EBL+) was obtained from Dr Balaguer (INSERM U1194, Montpellier, France). pGEX-RIP(27–439), (428–739), (683–1158)19 and pEFcmyc-RIP14032 were previously described. Platinum Select Human MLP Retroviral ShRNA-mir vectors were purchased from Ozyme (Montigny-le-Bretonneux, France).
LCoR was PCR amplified from MCF7 cell and cloned in pcDNA3.1(-) (Invitrogen, Cergy-Pontoise, France) to create pcDNA-LCoR. pEGFP-RIP140 (a kind gift of Dr Johanna Zilliacus, Karolinska Institutet, Huddinge, Sweden) was cloned in the BglII site of pCherry33 to create pCherry-RIP140. All LCoR fragments were PCR amplified, subcloned in p3XFLAG-myc-CMV (Sigma, Saint-Quentin, France), pECFP (Clontech, Mountain View, CA, USA), pM (Clontech), pGEX (Pharmacia, Uppsala, Sweden), pSG5-Puro or pcDNA3.1(-) (Invitrogen, Carlsbad, CA, USA) and sequenced. p1-Luc was constructed by PCR amplification of the LCoR promoter followed by cloning in the BglII and NCoI sites of pGL3-Basic (Promega).
Cell culture, transfections and luciferase assays
MCF7, T47D and COS7 cells were cultured in Ham’s F12/Dulbecco’s modified Eagle’s medium (1:1), RPMI or Dulbecco’s modified Eagle’s medium, respectively, supplemented with 10% fetal calf serum (Invitrogen) and antibiotics (Gibco, Illkirch, France). All cell lines were authenticated by short tandem repeat profiling and tested for mycoplasma contamination. For mRNA quantification, plasmids and siRNAs were transfected in MCF7 cells using JetPEI or Interferrin (Polyplus, Illkirch, France), respectively. For luciferase assays, MCF7 and COS7 cells were plated in 96-well plates and transfected with JetPEI (Polyplus) according to the manufacturer’s protocol. MEFs were cultured in F12/Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum and transfected using the calcium phosphate method. Transfection data were normalized by the renilla activity and expressed as relative luciferase activity. For immunoprecipitation assays, COS7 cells were plated at a density of 1.3 × 106 cells in 100 mm tissue culture dishes, treated, when indicated, with 100nM E2, and then harvested 18 h later. Protein expression was analyzed by western blotting.
To generate T47D cell lines that stably express LCoR variants, psg5-puromycin vectors expressing the different LCoR constructs were transfected by electroporation using the GenePulser Xcell (Bio-Rad, Marnes-la-Coquette, France), according to the manufacturer’s protocol. Cells were selected with 0.5 μg/ml puromycin.
Cell proliferation assay
T47D cells were seeded at a density of 2000 cells per well and MEFs at a density of 500 cells per well. At the indicated time, 0.5 mg/ml MTT (Sigma-Aldrich, St Louis, MO, USA) was added to selected wells and cells were incubated at 37 °C for 4h. Formazan crystals were solubilized in dimethylsulfoxide and absorbance read at 560 nm on a spectrophotometer. Results were normalized to the cell density at day 1.
GST pull-down assays
GST pull-down assays were performed as previously described.34 Detection of radioactivity was performed using a Fujix BAS5000 phosphoimager apparatus (Fujifilm, Marne-la-Vallée, France).
mRNA quantification
Breast cancer cell lines, normal breast tissues and primary breast cancer biopsies were collected in four French cancer hospitals: Institut regional du Cancer de Montpellier (Montpellier, France), Centre Léon Bérard (Lyon, France), Institut Paoli-Calmettes (Marseille, France) and Institut Gustave Roussy (Villejuif, France). RNA was then isolated using the Nucleospin RNA II Kit (Macherey-Nagel, Hoerdt, France) according to the manufacturer’s protocol. Reverse transcription–quantitative PCR assays were described elsewhere.14 The nucleotide sequences of the primers used for real-time PCR were: LCoR-F (5′-CATGTGCAGTATGGCTCGGA-3′), LCoR-R (5′-CATGCAGGCTCTACAGGCAAG-3′), LCoR2-F (5′-GAAATTAATGAGTGGATGACAATCGA-3′) and LCoR2-R (5′-CAGCCTACAATCTGAAATGACACAA-3′). A plate containing two identical sets of 48 tissue samples (four breast cancer stages and five normal tissues) (TissueScan Breast Cancer Tissue qPCR Panel II, Origene, Rockville, ML, USA) was analyzed by real-time PCR with an ABI Prism 7300 thermocycler (Ellange, Luxembourg).
Immunoprecipitation and immunoblotting assays
Immunoprecipitation and western blot assays were done as previously described35 using either anti-FLAG M2 Affinity Gel (Sigma), anti-c-Myc (sc-40, Santa Cruz, Dallas, TX, USA) or anti-Gal4DBD (sc-577, Santa Cruz) antibodies. Western blot analyses were performed using anti-FLAG (Sigma) or anti-c-Myc (sc-40, Santa Cruz) antibodies.
Proximity ligation assay
The Duolink II Proximity Ligation Assay kit (Eurogentec, Angers, France) was used according to the manufacturer’s instructions (Olink, Uppsala, Sweden). Primary antibodies were against RIP140 (Santa Cruz; sc-9459) and LCoR (Abcam, Cambridge, UK; ab48339). Signal detection was carried out by red fluorescence imaging performed using a Carl Zeiss Axioplan 2 (Marly-le-Roi, France) imaging microscope equipped with a 40 × objective.
Microscopy and image analysis
Images of transfected MCF7 and T47D cells were taken with an AxioImager Z2 (Carl Zeiss) using a 63 × 1.4N objective. scMOS Zyla 5.5 camera (Andor, Belfast, UK) was used for image acquisition and Image J (NIH) for image analysis. Transfected COS7 cells images were taken with an LSM510 META confocal microscope (Carl Zeiss) using a 40 × apochromat water immersion objective (numerical aperture, 1.2). Samples were sequentially illuminated at 488 and 543 nm, and subsequent fluorescence was detected using 505-530 BP and 560LP filters, respectively. Confocal microscopy software (SP2 version 3.2; Carl Zeiss MicroImaging, Inc., Jena, Germany) was used for capturing images, which were then analyzed by Image J (NIH, Bethesda, MD, USA).
Immunofluorescence analyses
Immunofluorescence assays were performed as previously described14 using an anti-LCoR antibody (ab48339, Abcam).
Breast tumor samples analysis
The Breast Cancer Gene-Expression Miner v3 statistical mining module (targeted correlation analysis)22 was used to determine the correlation between RIP140 and LCoR expression. The BreastMark algorithm (http://glados.ucd.ie/BreastMark) was used to perform Kaplan–Meier analysis on several data sets using either a median or a low cut-off.36 The PRECOG database37 was used to reanalyze the GSE24450 data set using a median cut-off and taking into account any event (overall survival and disease-specific survival).
Statistical analysis
All experiments were conducted independently at least three times. Results were expressed as the mean±s.d. Comparisons of two independent groups were performed using the Mann–Whitney U-test. A probability level (P-value) of 0.05 was chosen for statistical significance. Statistical analyses were performed using GraphPad Prism 5, version 5.01 (GraphPad Software, San Diego, CA, USA).
Acknowledgments
We thank Dr Arnaud Besson (CRCT, Toulouse, France) for providing MEF cells expressing or not p21, Dylane Detilleux for her help in characterizing stably transfected T47D cell lines. TNA received grants from Ligue Nationale contre le Cancer and Association pour la Recherche contre le Cancer. This work was funded by INSERM, Université de Montpellier and the Prix Ruban Rose.
Footnotes
Supplementary Information accompanies this paper on the Oncogene website (http://www.nature.com/onc)
The authors declare no conflict of interest.
Supplementary Material
References
- Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer J Int Cancer 2015; 136: E359–E386. [DOI] [PubMed] [Google Scholar]
- Gronemeyer H, Gustafsson J-A, Laudet V. Principles for modulation of the nuclear receptor superfamily. Nat Rev Drug Discov 2004; 3: 950–964. [DOI] [PubMed] [Google Scholar]
- Augereau P, Badia E, Carascossa S, Castet A, Fritsch S, Harmand P-O et al. The nuclear receptor transcriptional coregulator RIP140. Nucl Recept Signal 2006; 4: e024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes I, Bastien Y, Wai T, Nygard K, Lin R, Cormier O et al. Ligand-dependent nuclear receptor corepressor LCoR functions by histone deacetylase-dependent and -independent mechanisms. Mol Cell 2003; 11: 139–150. [DOI] [PubMed] [Google Scholar]
- Palijan A, Fernandes I, Bastien Y, Tang L, Verway M, Kourelis M et al. Function of histone deacetylase 6 as a cofactor of nuclear receptor coregulator LCoR. J Biol Chem 2009; 284: 30264–30274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palijan A, Fernandes I, Verway M, Kourelis M, Bastien Y, Tavera-Mendoza LE et al. Ligand-dependent corepressor LCoR is an attenuator of progesterone-regulated gene expression. J Biol Chem 2009; 284: 30275–30287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderon MR, Verway M, An B-S, DiFeo A, Bismar TA, Ann DK et al. Ligand-dependent corepressor (LCoR) recruitment by Kruppel-like factor 6 (KLF6) regulates expression of the cyclin-dependent kinase inhibitor CDKN1A gene. J Biol Chem 2012; 287: 8662–8674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderon MR, Verway M, Benslama RO, Birlea M, Bouttier M, Dimitrov V et al. Ligand-dependent corepressor contributes to transcriptional repression by C2H2 zinc-finger transcription factor ZBRK1 through association with KRAB-associated protein-1. Nucleic Acids Res 2014; 42: 7012–7027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Scully K, Zhu X, Cai L, Zhang J, Prefontaine GG et al. Opposing LSD1 complexes function in developmental gene activation and repression programmes. Nature 2007; 446: 882–887. [DOI] [PubMed] [Google Scholar]
- Asim M, Hafeez BB, Siddiqui IA, Gerlach C, Patz M, Mukhtar H et al. Ligand-dependent corepressor acts as a novel androgen receptor corepressor, inhibits prostate cancer growth, and is functionally inactivated by the Src protein kinase. J Biol Chem 2011; 286: 37108–37117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y, Shan S, Zhang Y, Liu W, Ding W, Ren W et al. Ligand-dependent corepressor acts as a novel corepressor of thyroid hormone receptor and represses hepatic lipogenesis in mice. J Hepatol 2012; 56: 248–254. [DOI] [PubMed] [Google Scholar]
- Cavaillès V, Dauvois S, L’Horset F, Lopez G, Hoare S, Kushner PJ et al. Nuclear factor RIP140 modulates transcriptional activation by the estrogen receptor. EMBO J 1995; 14: 3741–3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augereau P, Badia E, Balaguer P, Carascossa S, Castet A, Jalaguier S et al. Negative regulation of hormone signaling by RIP140. J Steroid Biochem Mol Biol 2006; 102: 51–59. [DOI] [PubMed] [Google Scholar]
- Lapierre M, Bonnet S, Bascoul-Mollevi C, Ait-Arsa I, Jalaguier S, Del Rio M et al. RIP140 increases APC expression and controls intestinal homeostasis and tumorigenesis. J Clin Invest 2014; 124: 1899–1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nautiyal J, Steel JH, Mane MR, Oduwole O, Poliandri A, Alexi X et al. The transcriptional co-factor RIP140 regulates mammary gland development by promoting the generation of key mitogenic signals. Dev Camb Engl 2013; 140: 1079–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz MH, Chen X, Zhang Q, DeFrain C, Osland J, Luo Y et al. Suppressing NRIP1 inhibits growth of breast cancer cells in vitro and in vivo. Oncotarget 2015; 6: 39714–39724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei J-J, Peng R-J, Kuang B-H, Yuan Z-Y, Qin T, Liu W-S et al. NOP14 suppresses breast cancer progression by inhibiting NRIP1/Wnt/β-catenin pathway. Oncotarget 2015; 6: 25701–25714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christian M, Kiskinis E, Debevec D, Leonardsson G, White R, Parker MG. RIP140-targeted repression of gene expression in adipocytes. Mol Cell Biol 2005; 25: 9383–9391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castet A, Boulahtouf A, Versini G, Bonnet S, Augereau P, Vignon F et al. Multiple domains of the receptor-interacting protein 140 contribute to transcription inhibition. Nucleic Acids Res 2004; 32: 1957–1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlando S, Gallastegui E, Besson A, Abril G, Aligué R, Pujol MJ et al. p27Kip1 and p21Cip1 collaborate in the regulation of transcription by recruiting cyclin-Cdk complexes on the promoters of target genes. Nucleic Acids Res 2015; 43: 6860–6873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guedj M, Marisa L, de Reynies A, Orsetti B, Schiappa R, Bibeau F et al. A refined molecular taxonomy of breast cancer. Oncogene 2012; 31: 1196–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jézéquel P, Frénel J-S, Campion L, Guérin-Charbonnel C, Gouraud W, Ricolleau G et al. bc-GenExMiner 3.0: new mining module computes breast cancer gene expression correlation analyses. Database J Biol Databases Curation 2013; 2013: bas060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muranen TA, Greco D, Fagerholm R, Kilpivaara O, Kämpjärvi K, Aittomäki K et al. Breast tumors from CHEK2 1100delC-mutation carriers: genomic landscape and clinical implications. Breast Cancer Res BCR 2011; 13: R90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X-J, Seto E. Lysine acetylation: codified crosstalk with other posttranslational modifications. Mol Cell 2008; 31: 449–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madak-Erdogan Z, Charn T-H, Jiang Y, Liu ET, Katzenellenbogen JA, Katzenellenbogen BS. Integrative genomics of gene and metabolic regulation by estrogen receptors α and β, and their coregulators. Mol Syst Biol 2013; 9: 676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho P-C, Tsui Y-C, Feng X, Greaves DR, Wei L-N. NF-κB-mediated degradation of the coactivator RIP140 regulates inflammatory responses and contributes to endotoxin tolerance. Nat Immunol 2012; 13: 379–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L, Anantharaman V, Balaji S, Babu MM, Iyer LM. The many faces of the helix-turn-helix domain: transcription regulation and beyond. FEMS Microbiol Rev 2005; 29: 231–262. [DOI] [PubMed] [Google Scholar]
- Abbas T, Dutta A. p21 in cancer: intricate networks and multiple activities. Nat Rev Cancer 2009; 9: 400–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosell M, Jones MC, Parker MG. Role of nuclear receptor corepressor RIP140 in metabolic syndrome. Biochim Biophys Acta 2011; 1812: 919–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulard C, Rambaud J, Lavergne E, Jacquemetton J, Renoir J-M, Trédan O et al. Role of JMJD6 in breast tumourigenesis. PloS One 2015; 10: e0126181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augereau P, Badia E, Fuentes M, Rabenoelina F, Corniou M, Derocq D et al. Transcriptional regulation of the human NRIP1/RIP140 gene by estrogen is modulated by dioxin signalling. Mol Pharmacol 2006; 69: 1338–1346. [DOI] [PubMed] [Google Scholar]
- Carascossa S, Gobinet J, Georget V, Lucas A, Badia E, Castet A et al. Receptor-interacting protein 140 is a repressor of the androgen receptor activity. Mol Endocrinol (Baltimore, MD) 2006; 20: 1506–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savatier J, Jalaguier S, Ferguson ML, Cavaillès V, Royer CA. Estrogen receptor interactions and dynamics monitored in live cells by fluorescence cross-correlation spectroscopy. Biochemistry (Mosc) 2010; 49: 772–781. [DOI] [PubMed] [Google Scholar]
- Duong V, Boulle N, Daujat S, Chauvet J, Bonnet S, Neel H et al. Differential regulation of estrogen receptor alpha turnover and transactivation by Mdm2 and stress-inducing agents. Cancer Res 2007; 67: 5513–5521. [DOI] [PubMed] [Google Scholar]
- Nait Achour T, Sentis S, Teyssier C, Philippat A, Lucas A, Corbo L et al. Transcriptional repression of estrogen receptor α signaling by SENP2 in breast cancer cells. Mol Endocrinol (Baltimore, MD) 2014; 28: 183–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madden SF, Clarke C, Gaule P, Aherne ST, O’Donovan N, Clynes M et al. BreastMark: an integrated approach to mining publicly available transcriptomic datasets relating to breast cancer outcome. Breast Cancer Res BCR 2013; 15: R52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentles AJ, Newman AM, Liu CL, Bratman SV, Feng W, Kim D et al. The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat Med 2015; 21: 938–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.