Fig. 2.
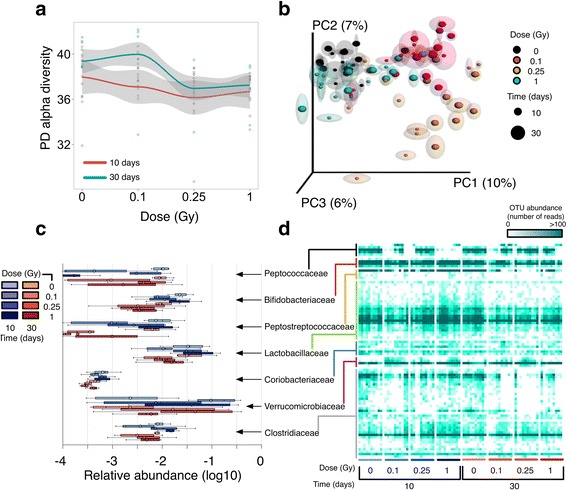
Ecological analysis of the irradiated microbiome. a Alpha diversity for control and irradiated samples 10 (red) and 30 (blue) days post-radiation. Shown are per-sample (dots), and per-condition averages (line plots), and standard deviations (gray bands). Values correspond to Faith’s phylogenetic diversity metric (PD). b Jackknifed Principal Coordinate Analysis (PCoA) plot of UniFrac unweighted distances between sample groups. For each sample, shown are confidence ellipses obtained from independent random rarefactions of the OTU counts table. c Barplots of per-condition relative abundances (logarithmic scale) for bacterial families with significant variations across conditions (Bonferroni p value < 0.05, Kruskal-Wallis test). d Heatmap of phylotype-level counts. All samples (columns) are shown and grouped by experimental factors. Individual phylotypes (rows) are grouped at the family level
