Fig. 6.
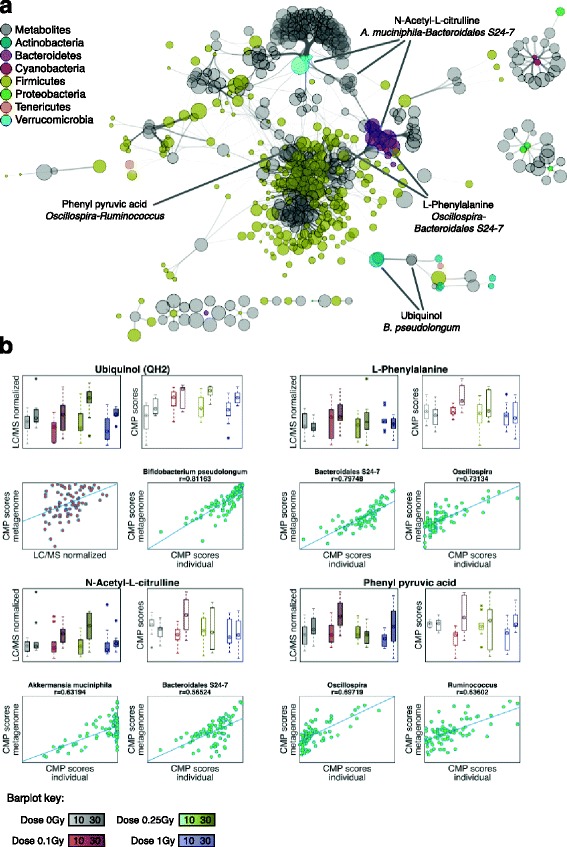
Metabolic network modeling and taxa-metabolite associations. Multi-omics (16S and LC-MS) data integration was performed under the Predicted Relative Metabolic Turnover (PRMT) framework. a Network visualization of significant associations between well-predicted metabolites (Mantel p value < 0.01 and FDR < 0.01, a total of 259 compounds) and bacterial phylotypes with a significant contribution to community-wide CMP scores (correlation between individual and community-wide CMP scores > 0.5 for a given metabolite, a total of 265 phylotypes). Node size is proportional to the relative abundance of the corresponding metabolite (from LC-MS) or phylotype (from 16S amplicon data). Edge width is proportional to the strength of association between each metabolite-phylotype pair (as measured by the correlation above). Highlighted are examples of well-predicted metabolites with significant agreement between experimental and predicted relative abundances and their association with specific phylotypes. b For each well-predicted metabolite highlighted in (a): solid barplots represent actual relative abundances (LC-MS); hollow barplots represent “predicted” relative abundances (CMP scores); red scatterplot for ubiquinol shows the correlation between actual and predicted relative abundances across all samples; green scatterplots show the correlation between community-wide and individual taxa contributions to predicted relative abundances, for taxa classified as key drivers of variations in metabolite relative abundances
