Figure 1.
PKC-3 Kinase Inhibition Leads to Symmetric Division and Loss of Asymmetry of Downstream Polarity Markers
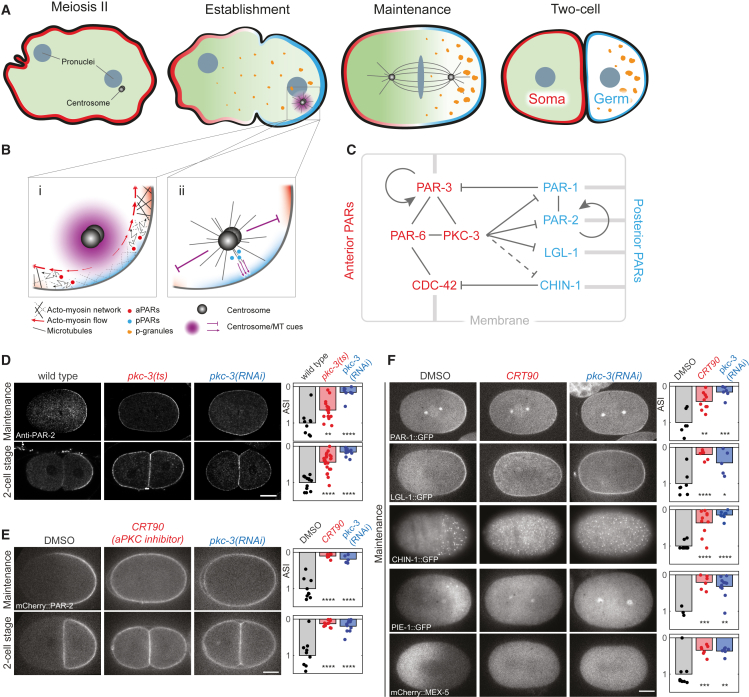
(A–C) Model for symmetry breaking by the PAR system in C. elegans. aPARs (red) initially occupy the membrane and pPARs (blue) are cytoplasmic (A, Meiosis II). A cue (purple) from the centrosome pair (black spheres) segregates aPARs into the anterior and promotes formation of a posterior PAR domain at the opposite pole (A, Establishment). PAR domains are then stable until cytokinesis (A, Maintenance) and drive polarization of cytoplasmic factors such as MEX-5/6 (green) and P granules (orange), which ensure the daughter cells acquire distinct fates (A, Two-cell). (B) Symmetry breaking can occur in two ways: (i) segregation of aPARs by cortical actomyosin flow (advection); and (ii) posterior PAR-2 loading. (C) A complex network of physical and regulatory interactions links the PAR proteins. Membrane binding (gray lines), physical interactions (black lines), as well as positive (→) and negative (⊥) feedback, are shown. Where links are indirect or unknown, dashed lines are used. Both CDC-42 and PAR-3 are required for stable membrane association of PAR-6/PKC-3. PAR-6 and PKC-3 depend on each other for membrane association. PAR-2, LGL-1, and presumably CHIN-1, are able to load onto the membrane independently. PAR-1 also binds membrane but requires PAR-2 to reach maximal concentrations. PKC-3 phosphorylates PAR-1, PAR-2, and LGL-1 and displaces them from the membrane. Exclusion of CHIN-1 from the anterior is dependent on PKC-3, but whether it is a direct target of PKC-3 is unknown. Together, PAR-1, via phosphorylation of PAR-3, and CHIN-1, by suppressing activated CDC-42, prevent invasion of the posterior domain by aPARs. PAR-3 and PAR-2 have been proposed to undergo oligomerization, which is thought to enhance their membrane association (noted by circular arrows). See recent reviews (Goehring, 2014, Hoege and Hyman, 2013, Motegi and Seydoux, 2013) for more information.
(D) Midsection confocal images of fixed zygotes stained for PAR-2 at polarity maintenance and two-cell stage comparing wild-type, pkc-3(ts), and pkc-3(RNAi) conditions.
(E) Midsection fluorescent images of mCherry:PAR-2-expressing zygotes at maintenance and two-cell stage in DMSO (control), CRT90-treated, and pkc-3(RNAi).
(F) Midsection (PAR-1, LGL-1, PIE-1, MEX-5) or cortical (CHIN-1) fluorescent images of maintenance-phase zygotes expressing markers to various downstream polarity markers in DMSO (control), CRT90-treated, and pkc-3(RNAi). Asymmetry in (D) to (F) is quantified by the asymmetry index, with one being normal asymmetry and zero, no asymmetry (ASI, normalized to DMSO/WT controls).
∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001. Scale bars, 10 μm. See also Figures S1 and S2; Movie S1.