Figure 2.
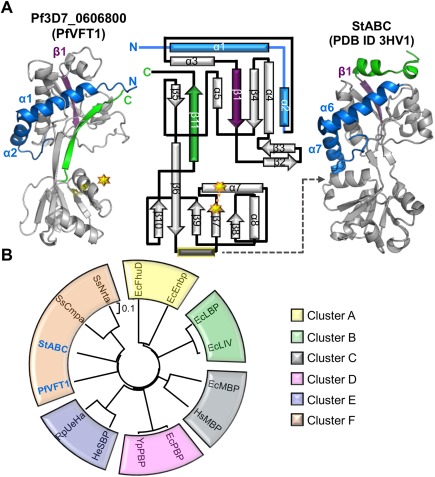
Pf3D7_0606800 adopts a Venus Flytrap (VFT) domain fold and is re‐annotated as PfVFT1. A: Secondary structure elements that are conserved (gray) and that diverge (purple, green, and blue) between PfVFT1 and StABC (PDB ID 3HV1). Topology diagram of PfVFT1 highlighting unique N‐terminus (blue components prior to β1, deep purple) and C‐terminus (green). Starburst and dotted connector indicate a disulfide bond. Gray dotted indicates lobe 2 helix (yellow outline) that is common to most VFTs, but absent in PfVFT1. B: A phylogenetic tree assembled using structure‐based (RMSD) alignments of PfVFT1 with two representative members for every cluster, obtained from previous evolutionary analysis on VFTs14 using PDBefold.15, 16 For simplicity, the recently identified Cluster G14 was not included. Escherichia coli (Ec); Homo sapiens (Hs); Yersinia pesits (Ys); Halomonas elongate (He); Ruegeria pomeroyi (Ru); Streptococcus thermophiles (St); Synechocystis spp. (Ss). EcFhuD‐1PSZ; EcFnbp‐2CHU; EcLBP‐1USK; EcLIV‐1Z16; EcMBP‐4MBP; HsMBP‐2ZVK; EcPBP‐1IXH, YsPBP‐2Z22; HeSBP‐2VPO; RuSBP‐3FXB; StABC‐3HV1; SsCmpa‐2I48; SsNrta‐2G29. An interactive view is available in the electronic version of the article.
