Figure 3.
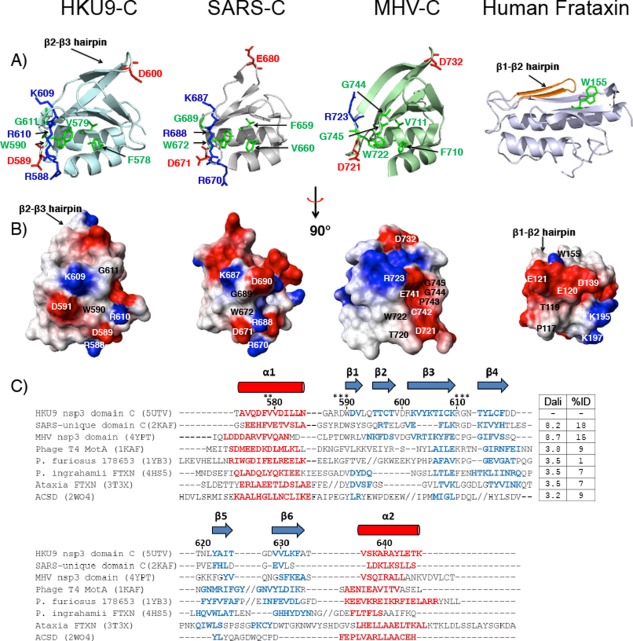
(A) Ribbon representations of nonstructural protein 3 C domains. Conserved residues or conservative substitutions relative to HKU9 are highlighted in green (aliphatic), blue (basic), and red (acidic). Residue numbers are indicated with respect to the first residue in each protein. Left to right: HKU9 C, SARS SUD‐C (PDB ID: 2KAF),35 MHV C domain (PDB ID: 4YPT),30 human frataxin (PDB ID: 3T3X).52 (B) C domain electrostatic potential surfaces. Red areas represent positively charged regions, blue areas represent negatively charged areas, and white areas represent neutral areas. (C) Structure‐based sequence alignment of the C domain in the Rousettus bat coronavirus HKU9, related viral proteins, and with other proteins in the frataxin fold family: phage T4 MotA (PDB ID: 1KAF),53 hypothetical protein (PDB ID: 1YB3), Psychromonas ingrahamii FTXN (PDB ID: 4HS5),52 Ataxia FTXN (PDB ID: 3T3X),54 AcsD (PDB ID: 2W04).55 The alignment is based on structural alignments obtained with TM‐Align.50 PDB codes are included after each protein name. The residue numbers for HKU9‐C are indicated. Alpha helix regions are displayed in red (cylinders) and beta strands are shown in blue (arrows). Gaps are shown as dashes (‐) and insertions where additional secondary structures are present are indicated by forward slash marks (//). Residues indicated by stars (*) discussed in the text are involved in potential functional sites. The corresponding Dali scores for the pairwise alignment of each protein with HKU9 C and the percent amino acid identity between each protein and HKU9 C domain are listed. Dali scores of 2.0 and higher indicate significant sequence identity51
