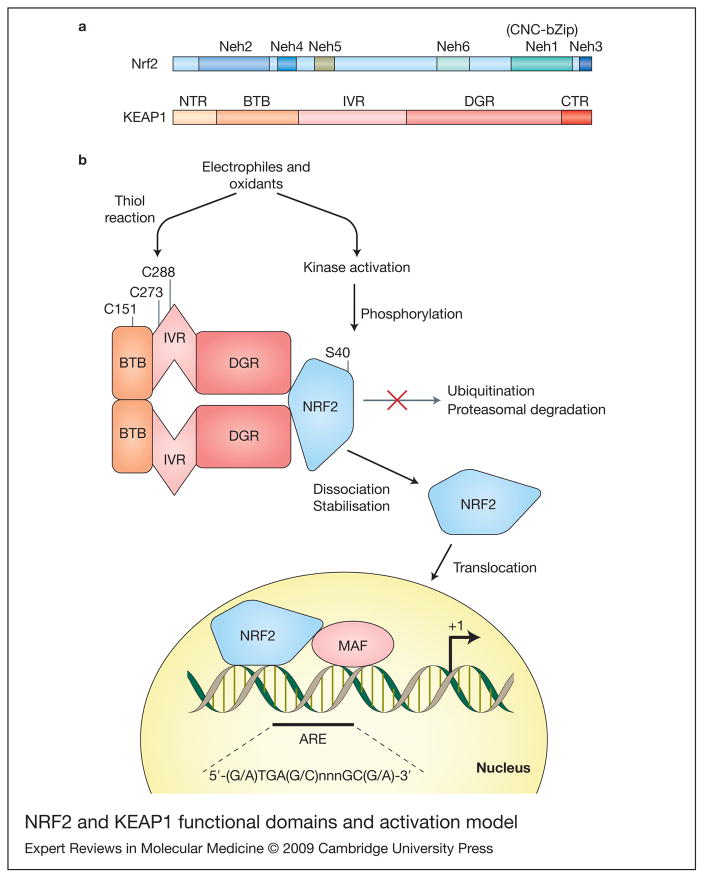
Figure 1. Nrf2 and KEAP1 functional domains and activation model.
(a) Diagram representing the functional domains of Nrf2 and Keap1. Nrf2 has six highly conserved regions named Neh1–Neh6 (Nrf2-ECH homology). The Neh1 contains the CNC homology region and basic-leucine zipper domain (CNC-bZip). The N-terminus (Neh2) and C-terminus (Neh3) of the proteins are also highly conserved. Keap1 binds Nrf2 at Neh2 and also the serine40 (S40) it is located in this domain. Additionally, there are two conserved acidic domains (Neh4 and Neh5) as well as a serine-rich conserved region (Neh6). KEAP1 presents two characteristic domains, the bricabrac, tramtrack and broad complex (BTB) domain and the double glycine repeat (DGR) domain. KEAP1 bridges the Cullin-3-based E3 ligase and Nrf2 using its BTB and the central intervening region (IVR) to bind Cul3 and its DGR to bind the Neh2 domain of Nrf2. Two additional regions are present in Nrf2: the N-terminal region (NTR) and the C-terminal region (CTR). (b) A model for KEAP1–Nrf2 interaction and activation. The BTB domain participates in KEAP1 dimerisation. Under basal conditions Nrf2 is continuously degraded. Electrophiles and oxidants directly modify reactive cysteines residues in KEAP1, disrupting its dimerisation or KEAP1–Cul3 interaction, and ubiquitination of Nrf2 is interrupted. Alternative activated kinases can phosphorylate Nrf2 at S40 and disrupt KEAP1–Nrf2 interaction. Nrf2 then translocates to the nucleus and increases ARE-driven transcription.