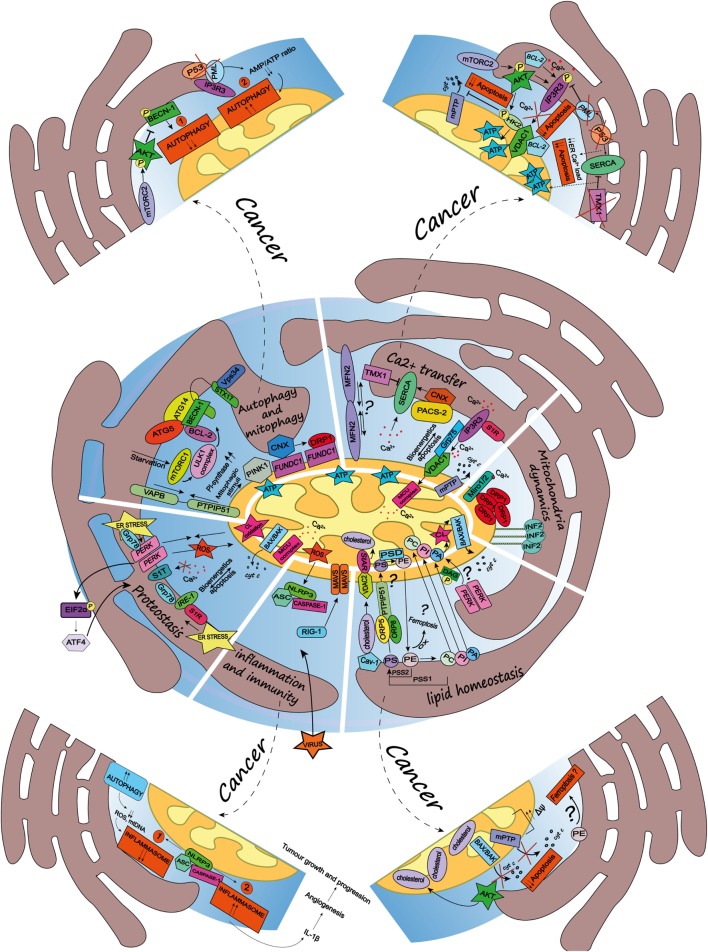
Figure 1.
Schematic summary of the multifaceted roles of the endoplasmic reticulum (ER)-mitochondria contact sites and representation of the main pathways they regulate. Here, are represented some of the vital cellular processes regulated by the mitochondria-associated membranes (MAMs), such as autophagy and mitophagy, Ca2+ transfer, mitochondria dynamics, lipid homeostasis, inflammation and immunity, and proteostasis. Dashed arrows link the physiological condition of some of the MAMs-regulated cellular functions to pathways harnessed by cancer (for further details see main text). Autophagy and mitophagy: here is shown the MAMs localization of the autophagy-related gene (ATG5) and ATG14, whose recruitment is mediated by the soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) protein syntaxin 17, upon activation of the mammalian target of rapamycin complex 1- Ulk1 unc-51 like kinase 1 (ULK1) complex, which is located in phosphatidylinositol synthase-enriched domains. Other autophagic key proteins found at MAMs are Beclin-1 and the phosphatidylinositol 3-kinase catalytic subunit type 3 (PIK3C3/Vps34). Mitophagic stimuli lead to the relocalization of the PTEN induced putative kinase 1 (PINK1) at MAMs. Additionally, the hypoxia-stimulated and mitochondria-associated FUN14 domain-containing 1, interacting with calnexin (CNX), relocates at MAMs, thus promoting Drp1 recruitment and hypoxia-induced mitophagy. Furthermore, the tethering complex vesicle-associated membrane protein-associated protein B (VAPB)–protein tyrosine phosphatase-interacting protein 51 (PTPIP51), which by tightening the ER–mitochondria contact sites, impairs rapamycin- and torin1-induced, but not starvation-induced, autophagy. Ca2+ transfer: here is represented the inositol 1,4,5 triphosphate receptor (IP3R) which is stabilized at MAMs by the chaperone Sigma1R (S1R). IP3R is involved in Ca2+ transfer from the ER storage to mitochondria by interacting with the mitochondrial voltage-dependent anion channel 1 (VDAC1), thus allowing Ca2+ transfer into the mitochondria through the mitochondrial calcium uniporter (MCU) and promoting mitochondrial bioenergetics and/or apoptosis, here represented by the opening of the mitochondrial permeability transition pore. Their interaction is allowed by the chaperone glucose-regulated protein 75, enriched at MAMs. Mitofusin 2 is another modulator of the interaction between ER and mitochondria (for details see the main text). The sarco/endoplasmic reticulum Ca2+ ATPase (SERCA) residing at the ER–mitochondria contact sites is positively modulated by chaperone CNX, whose localization at MAMs is regulated by the phosphofurin acidic cluster sorting protein 2 and negatively by the thioredoxin-related transmembrane protein 1. Mitochondria dynamics: here is represented the outer mitochondrial membrane (OMM) Rho-GTPase proteins Miro1/2 which, in presence of Ca2+, lose its binding with kinesin 1 (not shown), thereby reducing mitochondria motility and fostering the mitochondrial Ca2+ buffering. Oligomers formed by the dynamin-related guanosine triphosphatase (GTPase) dynamin-related protein 1 (Drp1) at MAMs, facilitate membrane scissions, along with actin filaments and the ER-localized protein inverted formin 2. Lipid homeostasis: the figure illustrates the lipid transfer and synthesis occurring at MAMs. The transfer of the phosphatidylserine (PS), which is formed from phosphatidylethanolamine (PE) or phosphatidylcholine (PC) by PS synthases (PSS1, PSS2), is required for the mitochondrial synthesis of PE, which is enabled by PS decarboxylase. Moreover, the ER-anchored oxysterol-binding protein-related protein (ORP) 5 and 8, are localized at MAMs where they could promote the transport of PS. PE, once exported to the ER is converted in PC. Here is also shown a speculative link to ferroptosis regulated at the MAMs, where the oxidation of polyunsaturated lipids, such as PE, occurs. The phosphatidic acid (PA) is transferred from the ER to the mitochondria as well, where it is essential for the synthesis of cardiolipin (CL), pivotal to apoptosis induction. Here is represented an interesting although speculative link involving the lipid activity kinase of the ER stress sensor RNA-dependent protein kinase (PKR)-like ER kinase (PERK), which by phosphorylating diacylglycerol can generate PA at MAMs. Cholesterol transfer into mitochondria is facilitated by the interaction with the cholesterol-binding caveolin-1, a MAM-resident protein. Moreover, the cholesterol transport steroidogenic acute regulatory protein can shuttle cholesterol from the OMM to the inner mitochondrial membrane interacting with the voltage-dependent anion channel 2. Inflammation and immunity: here is illustrated how the inflammasome is assembled at the MAMs after sensing host danger or intracellular damage, like damaged mitochondria. The inflammasome is made up of the pattern recognition receptors and nucleotide-binding domain and leucine-rich repeat-containing (NLR) protein family, NOD-like receptor family 3 and the adaptor protein apoptosis-associated speck-like protein (ASC), which recruits caspase-1 and allows its activation leading to the release of pro-inflammatory cytokines, like IL-1beta. Upon RNA virus infection, the recognition receptor RIG-I moved at MAMs, where it recruits its adaptor mitochondrial antiviral-signaling protein, thus starting an intracellular immune response. Proteostasis: here is reported that key ER stress sensors localize at MAMs, where, they can regulate signaling events linked to loss of ER homeostasis. In particular, the ER stress sensor PERK can modulate the tethering ER–mitochondria thus promoting the transfer of reactive oxygen species from the ER to the mitochondria, CL oxidation and the consequent BAX/BAK-dependent release of cytochrome c (cyt c). The PERK-activating transcription factor 4 axis of the unfolded protein response regulates the expression of S1T (truncated variants of SERCA1), thus leading to apoptosis. Moreover, here is also reported the MAMs localization of the inositol requiring enzyme 1, which interacts with S1R and Grp78, as well present at MAMs.