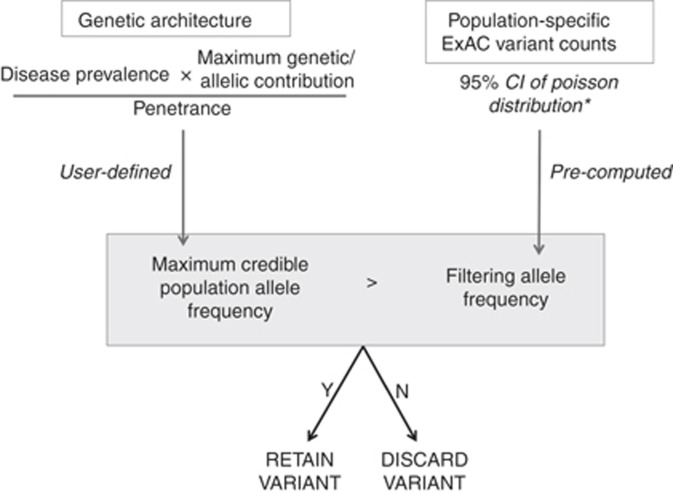
Figure 2.
A flow diagram of our approach, applied to a dominant condition, and using Exome Aggregation Consortium (ExAC) as our reference sample. First, a disease-level maximum credible population allele frequency (AF) is calculated, based on disease prevalence, heterogeneity, and penetrance. To evaluate a specific variant, we determine whether the observed variant allele count is compatible with disease by comparing this maximum credible population AF against the (precalculated) filtering AF for the variant. *While filtering AF has been precomputed for ExAC variants, the same framework can be readily applied using another reference sample.