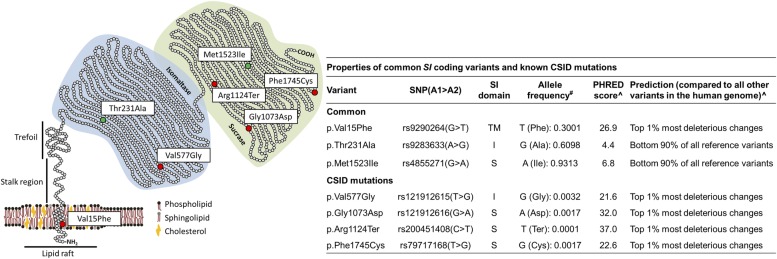
Figure 1.
Properties of sucrase–isomaltase (SI) mutants and common coding polymorphisms. Left: schematic representation of SI protein structure and functional domains; the position of congenital sucrase–isomaltase deficiency (CSID) mutations and common coding variants is reported and colour-coded according to their functional effects (red=damaging, green=benign). Right: the variants, corresponding dbSNP IDs (http://www.ncbi.nlm.nih.gov/SNP), SI protein domain location (TM, transmembrane; I, isomaltase; S, sucrase), allele frequency, PHRED-like score (range 1–99, ranking a variant relative to all possible substitutions in the human genome) and predicted functional consequences are reported. #Exome Aggregation Consortium browser (http://exac.broadinstitute.org); ^Combined Annotation-Dependent Depletion database (http://cadd.gs.washington.edu/info). SNP, single nucleotide polymorphism.