Figure 4.
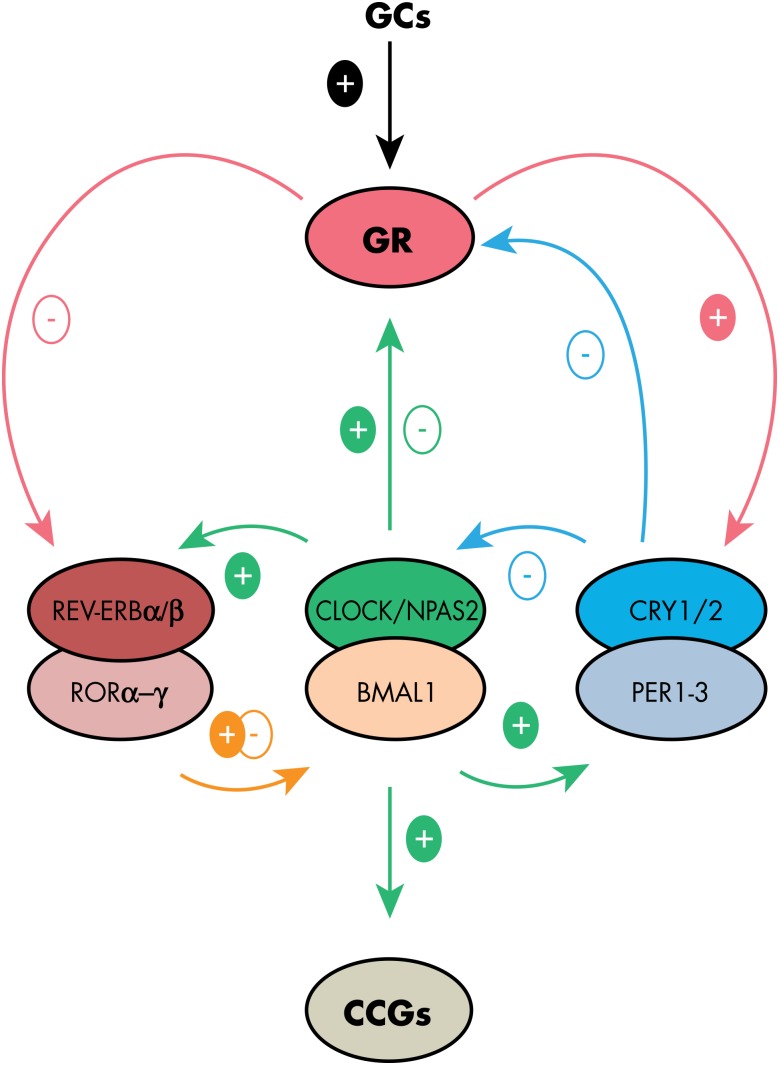
Molecular feedback loops generating circadian rhythmicity in both central and peripheral tissues. The upper part of the figure illustrates the interactions with GR activity. The basic transcription factors CLOCK (or its analog NPAS2) and BMAL1 heterodimerize and initiate the main positive loop by activation of other clock genes, including three Period genes (Per 1–3) and two Cryptochrome (Cry 1–2) genes. PER and CRY proteins form complexes that translocate to the nucleus where they inhibit their own CLOCK/BMAL1-induced transactivation, defining a main negative loop. CLOCK(NPAS2)/BMAL1 dimers also drive transcription of nuclear receptors of the REV-ERB and ROR families, including Rev-erb α-β (Nr1d1–2) and Ror α-β-γ. In turn, REVERBs and RORs inhibit and activate, respectively, the rhythmic transcription of Bmal1 and Clock. Although GR activation can reset the phase of the clock by regulating Per expression and REVERB activity, the clock machinery modulates GR activity at transcriptional and post-translational levels in multiple tissues, thus gating the regulation of GC target genes in a tissue-specific fashion.