ABSTRACT
Host factors, such as platelets, have been shown to enhance biofilm formation by oral commensal streptococci, inducing infective endocarditis (IE), but how bacterial components contribute to biofilm formation in vivo is still not clear. We demonstrated previously that an isogenic mutant strain of Streptococcus mutans deficient in autolysin AtlA (ΔatlA) showed a reduced ability to cause vegetation in a rat model of bacterial endocarditis. However, the role of AtlA in bacterial biofilm formation is unclear. In this study, confocal laser scanning microscopy analysis showed that extracellular DNA (eDNA) was embedded in S. mutans GS5 floes during biofilm formation on damaged heart valves, but an ΔatlA strain could not form bacterial aggregates. Semiquantification of eDNA by PCR with bacterial 16S rRNA primers demonstrated that the ΔatlA mutant strain produced dramatically less eDNA than the wild type. Similar results were observed with in vitro biofilm models. The addition of polyanethol sulfonate, a chemical lysis inhibitor, revealed that eDNA release mediated by bacterial cell lysis is required for biofilm initiation and maturation in the wild-type strain. Supplementation of cultures with calcium ions reduced wild-type growth but increased eDNA release and biofilm mass. The effect of calcium ions on biofilm formation was abolished in ΔatlA cultures and by the addition of polyanethol sulfonate. The VicK sensor, but not CiaH, was found to be required for the induction of eDNA release or the stimulation of biofilm formation by calcium ions. These data suggest that calcium ion-regulated AtlA maturation mediates the release of eDNA by S. mutans, which contributes to biofilm formation in infective endocarditis.
KEYWORDS: Streptococcus mutans, biofilm, extracellular DNA, infective endocarditis
INTRODUCTION
In addition to forming planktonic populations, bacteria can colonize and accumulate on surfaces to form biofilms, which are crucial in the pathogenesis of many subacute and chronic bacterial infections (1, 2). A typical example of a biofilm-associated disease is infective endocarditis (IE), which is most frequently caused by staphylococci or streptococci (3, 4). In particular, a population-based cohort study indicated that viridans group streptococci continue to outnumber Staphylococcus aureus as the most common organisms causing IE in the general population (5, 6). Our previous report showed that Streptococcus mutans and other commensal viridans group streptococci form biofilms on damaged heart valves. These biofilms were composed of layers of bacterial floes enclosed in activated platelet aggregates (7), but the detailed composition of the matrix inside the bacterial floes is not clear.
Bacterial biofilms are characterized by polymicrobial aggregates in the form of mats or floes and are typically encased in extracellular polymeric substances (3, 8, 9). Extracellular polymeric substances consist of a wide variety of polysaccharides, proteins, glycoproteins, glycolipids, and, in many cases, extracellular DNA (eDNA) (9–11). Although polysaccharides and proteins are important components, the role of eDNA as a critical element of the matrix, providing structural stability and protection against antimicrobial agents, has been increasingly recognized (12–15). eDNA was first shown to be present in the extracellular matrix of biofilms formed by Pseudomonas aeruginosa (16), but numerous studies since have reported similar eDNA properties in biofilms formed by a wide array of Gram-positive and -negative bacteria (14, 15, 17–19), suggesting that bacterial biofilm stabilization by eDNA is widespread.
eDNA release is typically a consequence of cell lysis. Bacterial murein hydrolases (bacterial autolysin) trigger autolytic cell wall digestion, leading to the release of DNA and other cellular contents into the extracellular environment (20). Bacterial autolysin has been implicated in biofilm formation, apparently by mediating bacterial lysis, with consequent release of eDNA, in diverse species, including Staphylococcus aureus, Staphylococcus epidermidis, and Enterococcus faecalis (14, 21, 22). Studies conducted on S. mutans have also demonstrated the roles of its autolysin protein AtlA in biofilm formation on polystyrene surfaces in vitro (23). In contrast to sucrose-mediated glucans, which are crucial for S. mutans biofilm formation on polystyrene and dental surfaces, the composition of intravascular S. mutans biofilm matrices is unclear. Therefore, the current study investigated the role of AtlA in S. mutans biofilm formation on the damaged heart valves of rats in a model of IE.
RESULTS
S. mutans eDNA mediates biofilm formation on damaged heart valves in vivo.
To address the role of eDNA in biofilm formation in IE, green fluorescent protein (GFP)-tagged S. mutans GS5 and atlA-deficient (ΔatlA) mutant strains were intravenously administered to rats with experimental IE. The vegetations were harvested and were analyzed by confocal laser scanning microscopy (CLSM). Detection of eDNA by staining with propidium iodide (PI) suggested that the eDNA was embedded inside S. mutans GS5 bacterial aggregates. In contrast, the ΔatlA strain appeared as single populations without embedded eDNA (Fig. 1A). To further confirm the existence of bacterial eDNA, total DNA was extracted from the harvested vegetations without breaking the bacteria. The eDNA was semiquantified by PCR with bacterial 16S rRNA primers. In agreement with the CLSM analysis, the level of eDNA detected in the ΔatlA sample was much lower than that in the parental wild-type strain (Fig. 1B and C). These data suggest that eDNA release mediated by AtlA contributes to S. mutans biofilm formation on damaged heart valves in rats with IE.
FIG 1.
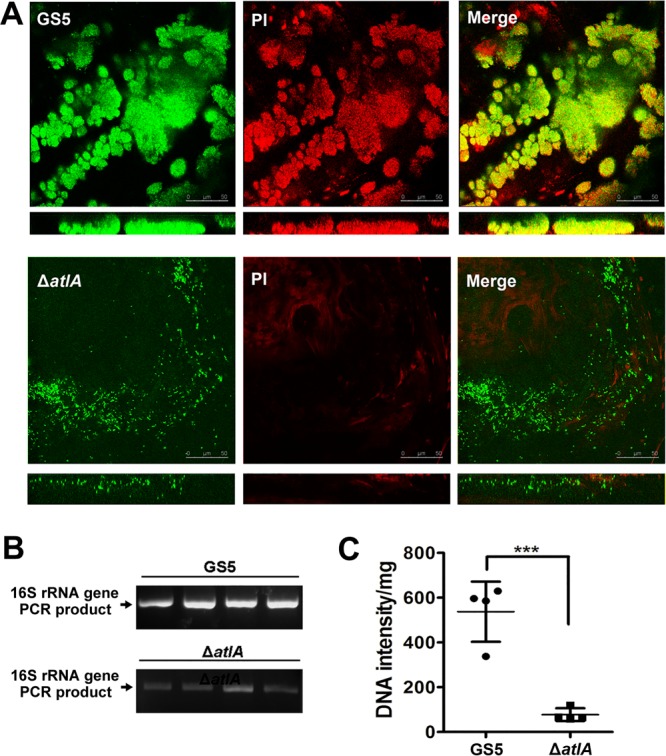
AtlA mediates eDNA release, contributing to S. mutans biofilm formation in vivo. (A) GFP-tagged wild-type (top) and ΔatlA (bottom) biofilms on vegetations harvested from injured rat heart valves were stained with 10 μM PI and were observed by CLSM (magnification, ×630). Yellow areas in merged images indicate the presence of both S. mutans and eDNA. These experiments were replicated three times, and results of a representative experiment are shown. (B and C) Total DNA isolated from vegetations (without breaking bacteria) was PCR amplified with bacterial 16S rRNA primers, and products were resolved on 1% agarose gels (B) and were quantified with ImageJ software (National Institutes of Health) (C). Data are means ± standard deviations from four experiments. Asterisks indicate significance (***, P ≤ 0.001) by Student's t test.
AtlA mediates cell lysis and the release of bacterial eDNA.
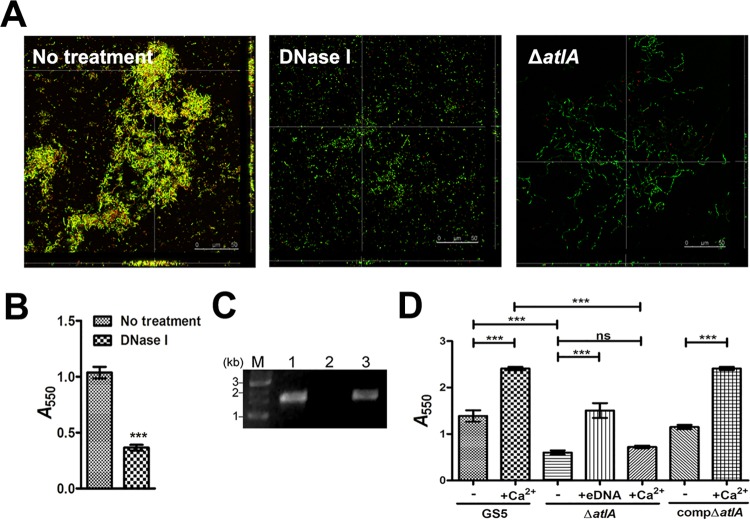
To confirm the role of bacterial eDNA, S. mutans biofilms were cultured in the defined culture medium without sucrose (24, 25). S. mutans biofilms formed aggregates in which eDNA was embedded, similar to those observed in vivo (Fig. 2A, left). Digestion of eDNA with DNase I dramatically reduced biofilm formation (Fig. 2A, center, and B), confirming the role of eDNA in S. mutans biofilm formation. In agreement with the in vivo data, the ΔatlA mutant strain exhibited a reduced ability to form biofilms and appeared as single populations without bacterial aggregates (Fig. 2A, right). Using bacterial 16S rRNA primers, we confirmed the lower level of eDNA release in the ΔatlA strain culture supernatant than in the wild-type and complemented (compΔatlA) strains (Fig. 2C). The addition of eDNA purified from the S. mutans GS5 culture supernatant restored the biofilm formation ability of the ΔatlA mutant strain (Fig. 2D), confirming the role of eDNA in S. mutans biofilm formation in the absence of sucrose.
FIG 2.
AtlA mediates bacterial eDNA release and biofilm formation. (A) CLSM images of GFP-tagged wild type S. mutans biofilms grown without (left) and with (center) DNase I and a ΔatlA biofilm (right) stained with 10 μM PI (magnification, ×630). Yellow areas indicate the presence of both S. mutans and eDNA. (B) Biofilms grown in 96-well plates with or without DNase I were stained with 0.1% crystal violet. Staining was detected by measuring the absorbance at 550 nm. Data are means ± standard deviations from triplicate experiments. Asterisks indicate significance (***, P < 0.001) by Student's t test. (C) eDNA released by wild-type (lane 1), ΔatlA (lane 2), and compΔatlA (lane 3) strains was amplified by PCR using bacterial 16S rRNA primers, and the products were resolved on 1% agarose gels. M, marker. (D) Biofilms of wild-type, ΔatlA, and compΔatlA strains were grown with or without purified bacterial eDNA or 0.1 mM calcium ions and were stained with 0.1% crystal violet. Staining was quantified by measuring the absorbance at 550 nm. Data are means ± standard deviations from triplicate experiments. Asterisks indicate significance (***, P ≤ 0.001) by one-way ANOVA. These experiments were repeated three times, and results of a representative experiment are shown.
eDNA is required for S. mutans biofilm adhesion and development.
To further address the roles of bacterial autolysis and eDNA in biofilm development, polyanethol sulfonate (PAS), a chemical lysis inhibitor, was used (26). The addition of PAS in the initial stages of biofilm formation abolished eDNA release into the culture medium and inhibited biofilm formation without affecting bacterial growth (Fig. 3A and B; Fig. 3C, 0-h time point). In addition, PAS treatment at the 2-, 4-, 6-, and 8-h time points resulted in dramatic reductions in the amount of adherent biomass; treatment after the biofilm was formed (24-h time point) had no effect (Fig. 3C). These data indicate that eDNA released by cell lysis is necessary at both the initial stage of biofilm adherence and the later stages of biofilm development.
FIG 3.
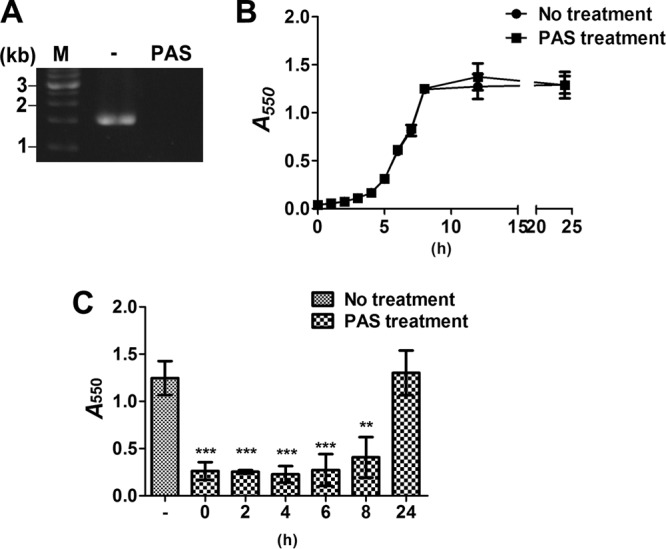
Inhibition of bacterial cell lysis by PAS reduces bacterial eDNA release and biofilm development. (A) eDNA released into the medium of S. mutans cultures with or without PAS was detected by PCR amplification using bacterial 16S rRNA primers and resolution of products on 1% agarose gels. M, Marker. (B) The growth curves of S. mutans cultured with or without PAS were measured by determining the optical density at 550 nm. (C) S. mutans cultures were treated with PAS at the time of inoculation (t = 0) and 2, 4, 6, 8, and 24 h postinoculation. All biofilms were grown at 37°C for a total of 24 h. Twenty-four-hour biofilms were allowed to grow for 3 h after the addition of PAS to allow full penetration and activity of the compound on the biofilm. The biofilms were stained with 0.1% crystal violet and were quantified by measuring the absorbance at 550 nm. Data are means ± standard deviations from triplicate experiments. Asterisks indicate significance (***, P ≤ 0.001; **, P ≤ 0.005) by one-way ANOVA. These experiments were repeated three times, and results of a representative experiment are shown.
Calcium ions enhance S. mutans eDNA release and biofilm formation.
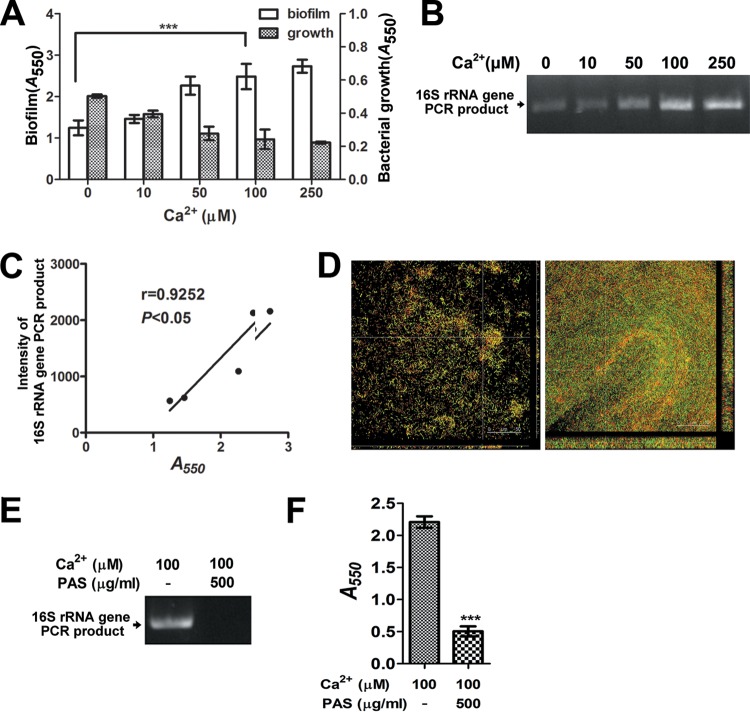
Our previous study reported that calcium ions in plasma can enhance the process of AtlA maturation, which modulates S. mutans autolytic activity (25). Therefore, the effects of calcium ions on eDNA release and biofilm formation were examined in the present study. The addition of various concentrations of calcium ions to the culture medium of S. mutans GS5 enhanced eDNA release and biofilm formation (Fig. 4A, B, and C). Interestingly, the addition of calcium ions also reduced bacterial growth in a dose-dependent manner (Fig. 4A). The enhancing effect of calcium ions on S. mutans biofilm formation was not observed with the ΔatlA strain but reappeared with the compΔatlA strain (Fig. 2D). CLSM imaging also showed that the addition of calcium ions enhanced the amount of eDNA embedded in bacterial biofilms and increased biofilm formation (Fig. 4D). The enhancement of eDNA release and biofilm formation by calcium ions was also consistently inhibited by the addition of PAS (Fig. 4E and F). Taken together, these data indicate that calcium ions enhance S. mutans cell lysis, which increases eDNA release and subsequent biofilm formation.
FIG 4.
Calcium ions enhance eDNA release and biofilm formation by S. mutans. (A) S. mutans biofilms grown with a variety of calcium ion concentrations were stained with 0.1% crystal violet and were quantified by measuring the absorbance at 550 nm. Overnight bacterial growth was also measured by determining the optical density at 550 nm. Data are means ± standard deviations from triplicate experiments. Asterisks indicate significance (***, P < 0.001) by one-way ANOVA. (B) eDNA released into the medium of S. mutans cultures without calcium ions or with different calcium ion concentrations was PCR amplified with bacterial 16S rRNA primers, and the products were resolved on 1% agarose gels. (C) The intensities of 16S rRNA gene PCR products were quantified by ImageJ software, and the correlation between DNA intensity and biofilms was further analyzed by a Pearson correlation test. (D) CLSM images represent S. mutans biofilms grown with (right) and without (left) 100 mM calcium (×630 magnification). S. mutans was labeled with GFP (green), and bacterial eDNA was stained with 10 μM PI. (E and F) S. mutans was cultured with 0.1 mM calcium, with or without PAS. (E) eDNA released into the medium was detected by PCR. (F) Biofilms were stained with 0.1% crystal violet and the absorbance quantified at 550 nm. Data are means ± standard deviations from triplicate experiments. Asterisks indicate significance (***. P < 0.001) by Student's t test. These experiments were repeated three times, and results of a representative experiment are shown.
VicK mediates calcium ion-enhanced AtlA maturation and biofilm formation.
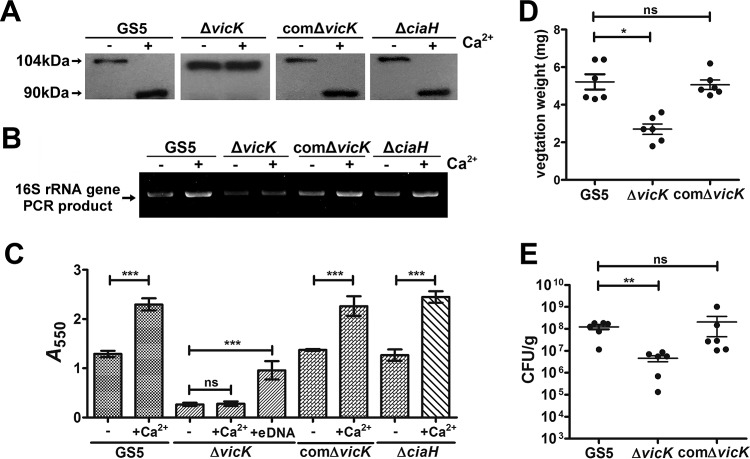
In S. mutans, 14 pairs of two-component systems (TCSs) have been identified by genomic analysis (27). Previous studies have reported that vicK disruption leads to inhibition of the N-terminal processing of AtlA (28) and that CiaH is responsible for calcium-mediated regulation in S. mutans (29). To investigate which TCS is responsible for sensing calcium ions and mediating eDNA release for biofilm formation, all of the isogenic mutants defective in any of the 14 TCSs were characterized except for the lethal mutant (data not shown). Consistently, the calcium ion-enhanced N-terminal processing of AtlA was abolished in a vicK-deficient (ΔvicK) mutant strain (Fig. 5A). The effects of calcium ions on eDNA release and biofilm formation were also abolished in the ΔvicK strain, but not in the ciaH-deficient (ΔciaH) strain, and the addition of bacterial eDNA to the culture medium enhanced biofilm formation by the ΔvicK strain (Fig. 5B and C). These results suggest a role for vicK in calcium ion sensing and in AtlA-mediated eDNA release and biofilm formation. Furthermore, the ΔvicK strain showed a reduced ability to colonize damaged heart valves in an experimental rat model of IE (Fig. 5D and E). Complementation with vicK restored all phenotypes of the ΔvicK strain (Fig. 5), confirming the role of vicK. These data suggest that VicK is responsible for sensing calcium ions in plasma and enhancing the ability of bacteria to form biofilms on damaged heart valves in IE in vivo.
FIG 5.
VicK, but not CiaH, is required for calcium ion sensing, induction of AtlA maturation, bacterial eDNA release, and enhanced biofilm formation. (A) S. mutans GS5, ΔvicK, comΔvicK, and ΔciaH strains were cultured in medium with or without the addition of calcium ions. Bacterial cell wall-associated proteins were isolated by use of 8 M urea and were subjected to Western blot analysis using anti-AtlA antibodies. Calcium ion-enhanced AtlA maturation was abolished in the vicK-deficient strain. (B) eDNA released into the medium of S. mutans GS5, ΔvicK, comΔvicK, and ΔciaH strains cultured with or without 100 μM calcium ions was PCR amplified using bacterial 16S rRNA primers, and products were resolved on 1% agarose gels. (C) The biofilms of S. mutans GS5, ΔvicK, comΔvicK, and ΔciaH strains grown in medium with or without purified bacterial eDNA and/or 0.1 mM calcium ions were stained with 0.1% crystal violet and were quantified by measuring the absorbance at 550 nm. Data are means ± standard deviations from triplicate experiments. Asterisks indicate significance (***, P < 0.001) by one-way ANOVA. These experiments were repeated three times, and results of a representative experiment are shown. (D and E) The role of VicK in the pathogenesis of S. mutans in IE was further investigated using the ΔvicK and comΔvicK strains in a rat model of IE. Vegetation size (D) and the numbers of colonized bacteria inside vegetations (E) were measured. Data represent means ± standard error of the means and were statistically analyzed using the Kruskal-Wallis test followed by Dunn's test (**, P < 0.01; *, P < 0.05).
DISCUSSION
The intravascular environment is very different from dental surfaces as a place in which to form bacterial biofilms, because it is filled with immune cells and extracellular matrix proteins but does not contain sucrose. Glucosyltransferase-deficient mutant strains, which cannot produce glucans, are still able to cause vegetation formation in the streptococcal IE model, suggesting a distinct matrix composition in S. mutans biofilms on the heart valve. While our previous study demonstrated that S. mutans biofilms that formed on damaged heart valves in vitro and in in vivo IE models were composed of bacterial floes enclosed in activated platelet aggregates (7), the matrix composition inside the bacterial floes remains unclear. The present study showed that the autolytic activity of AtlA mediated biofilm formation by S. mutans and the embedding of bacterial eDNA in the biofilms. In addition, we previously demonstrated the roles of neutrophil extracellular traps (NETs) in promoting vegetation maturation (30). In contrast to NETs, bacterial eDNA is embedded inside the bacterial aggregates and plays a role in connecting bacteria to each other, contributing to biofilm formation. The NETs formed inside the vegetation cover the bacterial biofilm and have the ability to induce thrombus formation, resulting in vegetation expansion and maturation (30). This may explain why DNase I treatment efficiently reduces both vegetation size and bacterial colony numbers in S. mutans-induced IE in rats (30).
In addition to biofilm formation, AtlA also play roles in modulating bacterial resistance to phagocytosis by binding to soluble fibronectin in the circulation (25). AtlA is able to bind various extracellular matrix proteins, including fibrinogen and collagen (data not shown), and may contribute to the ability of the bacteria to bind deposited platelets or exposed collagen on damaged heart valves. AtlA also contributes to S. mutans cell division and survival in the circulation (25). Therefore, AtlA plays multiple roles in the pathogenesis of S. mutans-induced IE.
Calcium-mediated N-terminal processing of AtlA results in its maturation, which enhances bacterial ability to bind soluble fibronectin in the circulation (25). It also enhances bacterial autolysis, which contributes to bacterial eDNA release and biofilm formation. More interestingly, enhanced bacterial autolysis results in increased bacterial death via “suicidal” or “fratricidal” mechanisms during biofilm development (21, 31). Although bacterial growth conditions would limit biofilm formation, our data showed that the intensity of the 16S rRNA gene PCR product is statistically correlated with biofilm formation (Fig. 4C), confirming that eDNA mediates biofilm formation. The N-terminal processing of AtlA is mediated by unknown bacterial protease machinery (25). The current results demonstrate that VicK plays a role in calcium ion-enhanced AtlA maturation and biofilm formation (Fig. 5). VicK has also been reported to have global roles in regulating S. mutans virulence, including acid tolerance, genetic competence, and oxidative stress tolerance (32–34). In addition, VicRK and CovR coordinate cell division and surface biogenesis with the extracellular synthesis of polysaccharides, a process required for the formation of structurally stable biofilms in the presence of sucrose (35). The current results also demonstrate the role of VicK in eDNA-dependent S. mutans biofilm formation in the absence of sucrose. Furthermore, the ΔvicK strain showed less ability to form biofilms in the medium without the addition of calcium ions than the wild-type strain (Fig. 5A), suggesting the involvement of other VicK-controlled factors in the pathogenesis of S. mutans-induced IE.
In summary, our current data demonstrate the role of VicK in sensing calcium ions in the circulation and regulating AtlA maturation, which contributes to eDNA release and bacterial biofilm formation on damaged heart valves in a rat model of S. mutans-induced IE. Since AtlA may be present in other viridans group streptococci and a homolog has also been found in other Gram-positive microorganisms, the results obtained from the present study provide important information regarding the pathogenesis of other forms of pathogen-induced IE.
MATERIALS AND METHODS
Bacterial strains and plasmid.
S. mutans GS5 and isogenic deletion mutant strains were grown and maintained in brain heart infusion broth (Difco Laboratories Inc., Detroit, MI, USA). The construction of the ΔatlA and compΔatlA strains is described in our previous study (25), and that of the ΔvicK and ΔciaH strains is described below. GFP-tagged bacteria were generated by transformation with a shuttle plasmid (pPDGFPuv) containing the GFPuv sequence described in our previous study (25). For the selection of antibiotic-resistant colonies after transformation, the growth medium was supplemented with chloramphenicol (5 μg/ml) (for the ΔatlA and compΔatlA strains), kanamycin (500 μg/ml) (for the ΔvicK, ΔciaH, comΔvicK, and compΔatlA strains), or spectinomycin (500 μg/ml) (for pPDGFPuv and the comΔvicK strain).
Constructions of the ΔvicK, ΔciaH, and comΔvicK strains.
All references to genomic loci are based on the S. mutans UA159 genome database (https://www.ncbi.nlm.nih.gov/genome/). To construct the ΔvicK and ΔciaH strains without the polar effect, these genes in S. mutans GS5 were disrupted by the insertion of a promoterless kanamycin cassette using a ligation-PCR mutagenesis strategy (36). Briefly, the promoterless kanamycin resistance gene fragment was isolated from the pALH124 plasmid by EcoRI digestion (37). Fragments of approximately 500 bp located before and/or after vicK and ciaH in the genome were PCR amplified using the forward and reverse primers listed in Table 1. PCR products were digested with EcoRI, ligated with the kanamycin resistance gene fragment, and transformed directly into S. mutans GS5. Correct allelic replacement in the derived isogenic mutant strains was confirmed by PCR amplification of the replacement region and reverse transcription-PCR (RT-PCR) analysis to verify the loss of vicK and ciaH expression and to confirm the expression of genes downstream of vicK and ciaH. For the construction of the comΔvicK strain, gene fragments of the vic operon promoter and vicK were PCR amplified using the primers listed in Table 1. PCR products were digested with BamHI and EcoRI and were cloned into the S. mutans–E. coli shuttle vector, PDL278, at the BamHI and EcoRI sites (25). The resulting plasmid was transformed into the ΔvicK strain to generate the comΔvicK strain. The expression of vicK was further confirmed by RT-PCR.
TABLE 1.
Primers for this study
| Primer | Sequence (5′ to 3′) | Target |
|---|---|---|
| VicKBF | AAGCTCTTGCCGTCTTT | vicK |
| VicKBR_EcoRI | GGAATTCCGGTGGGACTGGTTTAGGA | vicK |
| VicKAF_EcoRI | GGAATTCCGCCAGTAATCAAAGAACG | vicK |
| VicKAR | GTCTTGAAATTGGTGCT | vicK |
| CiaHBF | AACGTGCTGAACTTAACG | ciaH |
| CiaHBR_EcoRI | GGAATTCCGATAAAGCCAGAACACGA | ciaH |
| CiaHAF_EcoRI | GGAATTCCGTATTAGCAGCCATCTGG | ciaH |
| CiaHAR | AACTGCTTGGCAAAGAGT | ciaH |
| VicKPF_BamHI | CGCGGATCCGGTAATGAGTGCCATAGT | vic promoter |
| VicKPR_EcoRI | CCGGAATTCAATTTTCTTCATTATGAATACC | vic promoter |
| VicKF_EcoRI | CCGGAATTCATGACTAATGTGTTTGAATCA | vicK |
| VicKR_EcoRI | CCGGA TTCTGTTTCTGTCATGATTCG | vicK |
Biofilm formation assay.
To establish sucrose-independent biofilms, sucrose-free defined M4 medium was used. Bacterial biofilm growth was initiated by inoculating individual wells of a 96-well polystyrene microtiter plate with approximately 107 CFU in 200 μl of M4 medium. To investigate the roles of bacterial eDNA and cell lysis in biofilm formation, DNase I (2 U/ml) and PAS (500 μg/ml) were added to the medium, respectively. After a 16-h incubation at 37°C, the medium was removed and the wells rinsed gently three times with sterile distilled water. The plates were then air dried, stained with 0.1% crystal violet for 10 min, rinsed three times with distilled water, and air dried again for 10 min. Each assay was performed in triplicate, and wells without biofilms were used as blank controls. Biofilms were quantified by measuring the absorbance at 550 nm using a MicroELISA reader (Dynatech Corp., Alexandria, VA, USA).
For CLSM analysis, GFPuv-tagged strains were transformed with pPDGFPuv, and the bacterial biofilms were cultured in a 24-well plate with a round glass coverslip in individual wells. After a 16-h incubation, biofilms that had formed on the glass coverslip were gently washed three times with phosphate-buffered saline and then fixed with 2% paraformaldehyde for 15 min. After fixation, bacterial eDNA embedded in the biofilm was stained with 10 μM PI. After three washes with phosphate-buffered saline, the coverslips were transferred to a slide and were observed by CLSM (Leica TCS SP5 confocal microscope).
Purification and semiquantification of eDNA.
To detect eDNA release, the culture medium was collected and analyzed. Bacteria were cultured in M4 medium with or without PAS (500 μg/ml) and calcium ions. Before collection of the culture medium, biofilms adherent to the culture tubes were disrupted and suspended by violent agitation using a vortex. Bacterial cells were removed by centrifugation for 2 min at 16,000 × g andt 4°C. To specifically detect eDNA release, 1 μl of supernatant was used as a template for semiquantitative PCR (25 cycles) using specific bacterial 16S rRNA primers. M4 medium (1 μl) without seeding bacteria was used as a negative control, and medium containing S. mutans GS5 genomic DNA was used as a positive PCR control.
eDNA released into the culture medium was purified by collecting 0.5 ml of the culture medium supernatant and saturating it with a phenol-chloroform-isoamyl alcohol mix (25:24:1). After vortexing for 30 s, the mixture was centrifuged at 4°C for 5 min at 16,000 × g. The aqueous phase (0.3 ml) was removed and was mixed with 30 μl of 5 M NaCl and 800 μl of 100% ethanol. The mixture was first incubated at −80°C and then centrifuged for 10 min at 16,000 × g. The supernatant was decanted, and the precipitated sample was air dried and was suspended in 30 μl of double-distilled water. For the biofilm assay, 20 μl of purified eDNA was added to 200 μl of M4 medium. The total DNA of the vegetation was extracted using a Gentra Puregene tissue kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. The bacterial eDNA was detected by semiquantitative PCR (25 cycles) using specific bacterial 16S rRNA primers.
S. mutans-induced IE rat model.
Approval of animal use was obtained from the National Taiwan University Institutional Animal Care and Use Committee (Taipei, Taiwan, Republic of China) prior to initiation of the experiments. A modified rat model of experimental streptococcal endocarditis was used as described previously (38). For CLSM analysis, GFP-tagged wild-type S. mutans GS5 or its ΔatlA mutant (1 × 109 CFU) was injected into the tail veins of Wistar rats. The vegetation harvested from the heart of each rat was fixed on a glass slide and was stained with 10 μM PI. Bacterial biofilms were then observed under a confocal microscope (Leica TCS SP5).
Statistical analysis.
The statistical significance of the difference between two sets of data was analyzed by using an unpaired, two-tailed Student t test. Differences between more than two sets of data were assessed using one-way analysis of variance (ANOVA) followed by the Bonferroni multiple-comparison test. For nonparametrically distributed data, a Kruskal-Wallis test, followed by Dunn's test, was used. A P value of <0.05 was considered statistically significant.
ACKNOWLEDGMENTS
We declare that we have no conflicts of interest.
This study was supported by the Ministry of Science and Technology of Taiwan (MOST) (grants 103-2320-B-002-037-MY3, 103-2320-B-002-045-MY3, and 106-2320-B-038-004-MY2) and by the Academia Sinica and MOST (grants 104-0210-01-09-02, 105-0210-01-13-01, and 106-0210-01-15-02).
REFERENCES
- 1.Donlan RM, Costerton JW. 2002. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin Microbiol Rev 15:167–193. doi: 10.1128/CMR.15.2.167-193.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Parsek MR, Singh PK. 2003. Bacterial biofilms: an emerging link to disease pathogenesis. Annu Rev Microbiol 57:677–701. doi: 10.1146/annurev.micro.57.030502.090720. [DOI] [PubMed] [Google Scholar]
- 3.Hall-Stoodley L, Costerton JW, Stoodley P. 2004. Bacterial biofilms: from the natural environment to infectious diseases. Nat Rev Microbiol 2:95–108. doi: 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- 4.Moreillon P, Que YA. 2004. Infective endocarditis. Lancet 363:139–149. doi: 10.1016/S0140-6736(03)15266-X. [DOI] [PubMed] [Google Scholar]
- 5.Tleyjeh IM, Steckelberg JM, Murad HS, Anavekar NS, Ghomrawi HM, Mirzoyev Z, Moustafa SE, Hoskin TL, Mandrekar JN, Wilson WR, Baddour LM. 2005. Temporal trends in infective endocarditis: a population-based study in Olmsted County, Minnesota. JAMA 293:3022–3028. doi: 10.1001/jama.293.24.3022. [DOI] [PubMed] [Google Scholar]
- 6.Fowler VG Jr, Miro JM, Hoen B, Cabell CH, Abrutyn E, Rubinstein E, Corey GR, Spelman D, Bradley SF, Barsic B, Pappas PA, Anstrom KJ, Wray D, Fortes CQ, Anguera I, Athan E, Jones P, van der Meer JT, Elliott TS, Levine DP, Bayer AS, ICE Investigators . 2005. Staphylococcus aureus endocarditis: a consequence of medical progress. JAMA 293:3012–3021. doi: 10.1001/jama.293.24.3012. [DOI] [PubMed] [Google Scholar]
- 7.Jung CJ, Yeh CY, Shun CT, Hsu RB, Cheng HW, Lin CS, Chia JS. 2012. Platelets enhance biofilm formation and resistance of endocarditis-inducing streptococci on the injured heart valve. J Infect Dis 205:1066–1075. doi: 10.1093/infdis/jis021. [DOI] [PubMed] [Google Scholar]
- 8.Costerton JW, Cheng KJ, Geesey GG, Ladd TI, Nickel JC, Dasgupta M, Marrie TJ. 1987. Bacterial biofilms in nature and disease. Annu Rev Microbiol 41:435–464. doi: 10.1146/annurev.mi.41.100187.002251. [DOI] [PubMed] [Google Scholar]
- 9.Flemming HC, Neu TR, Wozniak DJ. 2007. The EPS matrix: the “house of biofilm cells.” J Bacteriol 189:7945–7947. doi: 10.1128/JB.00858-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goller CC, Romeo T. 2008. Environmental influences on biofilm development. Curr Top Microbiol Immunol 322:37–66. [DOI] [PubMed] [Google Scholar]
- 11.Karatan E, Watnick P. 2009. Signals, regulatory networks, and materials that build and break bacterial biofilms. Microbiol Mol Biol Rev 73:310–347. doi: 10.1128/MMBR.00041-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mulcahy H, Charron-Mazenod L, Lewenza S. 2008. Extracellular DNA chelates cations and induces antibiotic resistance in Pseudomonas aeruginosa biofilms. PLoS Pathog 4:e1000213. doi: 10.1371/journal.ppat.1000213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Petersen FC, Tao L, Scheie AA. 2005. DNA binding-uptake system: a link between cell-to-cell communication and biofilm formation. J Bacteriol 187:4392–4400. doi: 10.1128/JB.187.13.4392-4400.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qin Z, Ou Y, Yang L, Zhu Y, Tolker-Nielsen T, Molin S, Qu D. 2007. Role of autolysin-mediated DNA release in biofilm formation of Staphylococcus epidermidis. Microbiology 153:2083–2092. doi: 10.1099/mic.0.2007/006031-0. [DOI] [PubMed] [Google Scholar]
- 15.Vilain S, Pretorius JM, Theron J, Brozel VS. 2009. DNA as an adhesin: Bacillus cereus requires extracellular DNA to form biofilms. Appl Environ Microbiol 75:2861–2868. doi: 10.1128/AEM.01317-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Whitchurch CB, Tolker-Nielsen T, Ragas PC, Mattick JS. 2002. Extracellular DNA required for bacterial biofilm formation. Science 295:1487. doi: 10.1126/science.295.5559.1487. [DOI] [PubMed] [Google Scholar]
- 17.Mann EE, Rice KC, Boles BR, Endres JL, Ranjit D, Chandramohan L, Tsang LH, Smeltzer MS, Horswill AR, Bayles KW. 2009. Modulation of eDNA release and degradation affects Staphylococcus aureus biofilm maturation. PLoS One 4:e5822. doi: 10.1371/journal.pone.0005822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lappann M, Claus H, van Alen T, Harmsen M, Elias J, Molin S, Vogel U. 2010. A dual role of extracellular DNA during biofilm formation of Neisseria meningitidis. Mol Microbiol 75:1355–1371. doi: 10.1111/j.1365-2958.2010.07054.x. [DOI] [PubMed] [Google Scholar]
- 19.Spoering AL, Gilmore MS. 2006. Quorum sensing and DNA release in bacterial biofilms. Curr Opin Microbiol 9:133–137. doi: 10.1016/j.mib.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 20.Rice KC, Bayles KW. 2008. Molecular control of bacterial death and lysis. Microbiol Mol Biol Rev 72:85–109. doi: 10.1128/MMBR.00030-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thomas VC, Hiromasa Y, Harms N, Thurlow L, Tomich J, Hancock LE. 2009. A fratricidal mechanism is responsible for eDNA release and contributes to biofilm development of Enterococcus faecalis. Mol Microbiol 72:1022–1036. doi: 10.1111/j.1365-2958.2009.06703.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rice KC, Mann EE, Endres JL, Weiss EC, Cassat JE, Smeltzer MS, Bayles KW. 2007. The cidA murein hydrolase regulator contributes to DNA release and biofilm development in Staphylococcus aureus. Proc Natl Acad Sci U S A 104:8113–8118. doi: 10.1073/pnas.0610226104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shibata Y, Kawada M, Nakano Y, Toyoshima K, Yamashita Y. 2005. Identification and characterization of an autolysin-encoding gene of Streptococcus mutans. Infect Immun 73:3512–3520. doi: 10.1128/IAI.73.6.3512-3520.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takada K, Fukushima K. 1986. Effects of certain salts on glucosyltransferase synthesis by Streptococcus mutans strain PS-14. J Dent Res 65:452–455. doi: 10.1177/00220345860650031601. [DOI] [PubMed] [Google Scholar]
- 25.Jung CJ, Zheng QH, Shieh YH, Lin CS, Chia JS. 2009. Streptococcus mutans autolysin AtlA is a fibronectin-binding protein and contributes to bacterial survival in the bloodstream and virulence for infective endocarditis. Mol Microbiol 74:888–902. doi: 10.1111/j.1365-2958.2009.06903.x. [DOI] [PubMed] [Google Scholar]
- 26.Wecke J, Lahav M, Ginsburg I, Kwa E, Giesbrecht P. 1986. Inhibition of wall autolysis of staphylococci by sodium polyanethole sulfonate “liquoid.” Arch Microbiol 144:110–115. [DOI] [PubMed] [Google Scholar]
- 27.Biswas I, Drake L, Erkina D, Biswas S. 2008. Involvement of sensor kinases in the stress tolerance response of Streptococcus mutans. J Bacteriol 190:68–77. doi: 10.1128/JB.00990-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ahn SJ, Burne RA. 2007. Effects of oxygen on biofilm formation and the AtlA autolysin of Streptococcus mutans. J Bacteriol 189:6293–6302. doi: 10.1128/JB.00546-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.He X, Wu C, Yarbrough D, Sim L, Niu G, Merritt J, Shi W, Qi F. 2008. The cia operon of Streptococcus mutans encodes a unique component required for calcium-mediated autoregulation. Mol Microbiol 70:112–126. doi: 10.1111/j.1365-2958.2008.06390.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jung CJ, Yeh CY, Hsu RB, Lee CM, Shun CT, Chia JS. 2015. Endocarditis pathogen promotes vegetation formation by inducing intravascular neutrophil extracellular traps through activated platelets. Circulation 131:571–581. doi: 10.1161/CIRCULATIONAHA.114.011432. [DOI] [PubMed] [Google Scholar]
- 31.Thomas VC, Hancock LE. 2009. Suicide and fratricide in bacterial biofilms. Int J Artif Organs 32:537–544. [DOI] [PubMed] [Google Scholar]
- 32.Senadheera D, Krastel K, Mair R, Persadmehr A, Abranches J, Burne RA, Cvitkovitch DG. 2009. Inactivation of VicK affects acid production and acid survival of Streptococcus mutans. J Bacteriol 191:6415–6424. doi: 10.1128/JB.00793-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Senadheera MD, Guggenheim B, Spatafora GA, Huang YC, Choi J, Hung DC, Treglown JS, Goodman SD, Ellen RP, Cvitkovitch DG. 2005. A VicRK signal transduction system in Streptococcus mutans affects gtfBCD, gbpB, and ftf expression, biofilm formation, and genetic competence development. J Bacteriol 187:4064–4076. doi: 10.1128/JB.187.12.4064-4076.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Deng DM, Liu MJ, ten Cate JM, Crielaard W. 2007. The VicRK system of Streptococcus mutans responds to oxidative stress. J Dent Res 86:606–610. doi: 10.1177/154405910708600705. [DOI] [PubMed] [Google Scholar]
- 35.Stipp RN, Boisvert H, Smith DJ, Hofling JF, Duncan MJ, Mattos-Graner RO. 2013. CovR and VicRK regulate cell surface biogenesis genes required for biofilm formation in Streptococcus mutans. PLoS One 8:e58271. doi: 10.1371/journal.pone.0058271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lau PC, Sung CK, Lee JH, Morrison DA, Cvitkovitch DG. 2002. PCR ligation mutagenesis in transformable streptococci: application and efficiency. J Microbiol Methods 49:193–205. doi: 10.1016/S0167-7012(01)00369-4. [DOI] [PubMed] [Google Scholar]
- 37.Kremer BH, van der Kraan M, Crowley PJ, Hamilton IR, Brady LJ, Bleiweis AS. 2001. Characterization of the sat operon in Streptococcus mutans: evidence for a role of Ffh in acid tolerance. J Bacteriol 183:2543–2552. doi: 10.1128/JB.183.8.2543-2552.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shun CT, Lu SY, Yeh CY, Chiang CP, Chia JS, Chen JY. 2005. Glucosyltransferases of viridans streptococci are modulins of interleukin-6 induction in infective endocarditis. Infect Immun 73:3261–3270. doi: 10.1128/IAI.73.6.3261-3270.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]