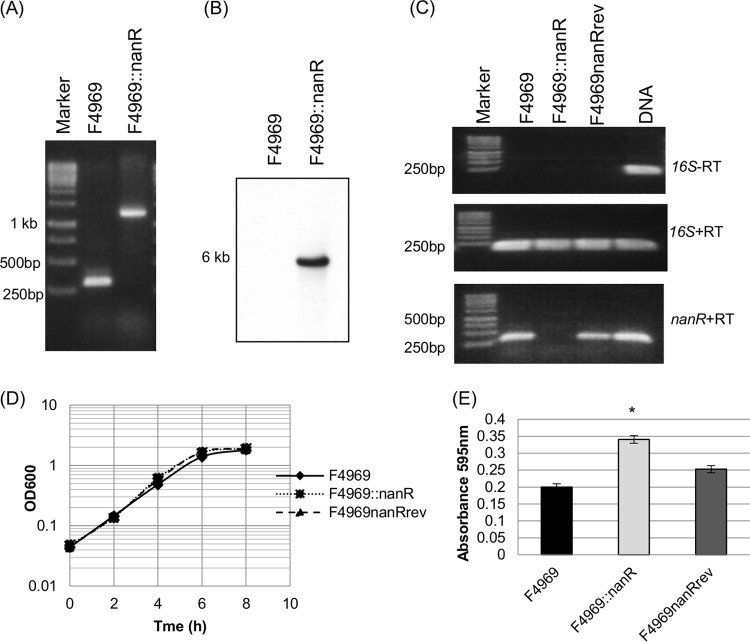
FIG 5.
Construction and characterization of an F4969 nanR null mutant and a reversed mutant strain. (A) PCR confirmation of isogenic nanR null mutant strain construction. Using DNA from F4969, a PCR using internal nanR gene primers amplified the expected product of ∼300 bp. Using template DNA isolated from the F4969::nanR strain, which has a 900-bp intron insertion in the nanR gene, the same PCR assay amplified a larger product, of ∼1,200 bp. The left lane shows a 1-kb molecular ruler. (B) Intron-specific Southern blot hybridization with DNA from the wild-type or nanR null mutant strain. The size of the DNA fragment is indicated at left. (C) RT-PCR analysis of nanR gene transcription in wild-type, null mutant, and reversed strains grown for 4 h at 30°C. (Top) Transcription analysis of the housekeeping 16S rRNA gene without reverse transcriptase (−RT) indicated that all RNAs were free of DNA contamination. (Middle) transcription analysis of the housekeeping 16S rRNA gene with reverse transcriptase (+RT) indicated that the isolated RNA was of high quality. (Bottom) Transcription analysis of the nanR gene. The left lane shows the 1-kb molecular ruler, and the right lane shows the F4969 DNA (as a positive control). (D) Postinoculation changes in OD600 for cultures of wild-type, null mutant, and reversed mutant strains grown in TH medium at 30°C. The experiment was repeated three times, and representative results are shown. (E) Supernatant sialidase activity analysis of wild-type, null mutant, and reversed mutant strains grown overnight in TH medium at 30°C. The experiment for panel E was repeated three times, and mean values are shown. The error bars indicate standard deviations. *, P < 0.05 compared to the wild-type strain by ordinary one-way ANOVA.