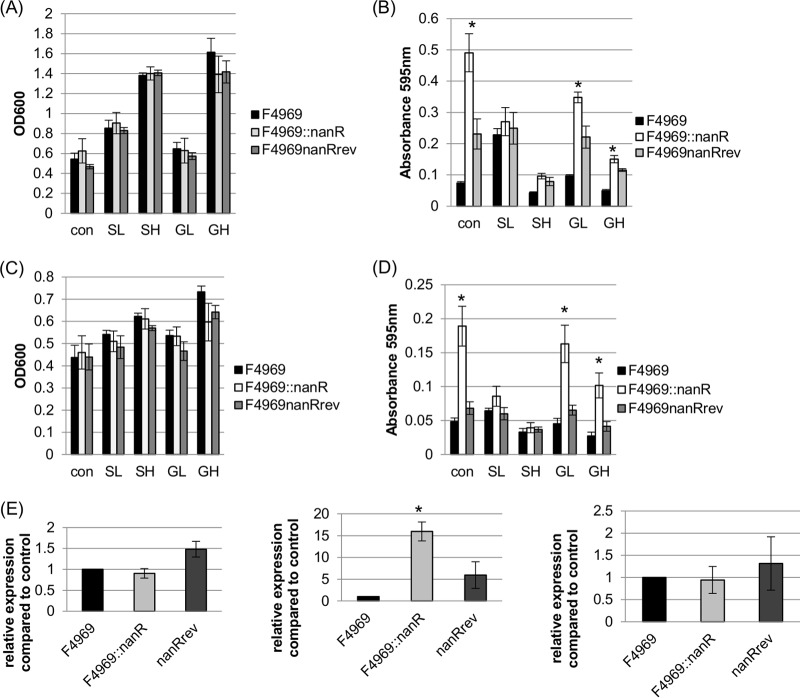
FIG 6.
Supernatant sialidase activities and nanI transcription levels of wild-type F4969, an isogenic nanR null mutant, and the reversed mutant cultured at 30°C in DMEM with or without glucose or sialic acid. (A) Postinoculation OD600 changes for 24-h cultures of the wild-type, nanR null mutant, and reversed mutant strains of F4969 grown in control DMEM, DMEM with 1.6 mM sialic acid (SL), DMEM with 1.6 mM glucose (GL), DMEM with 16 mM sialic acid (SH), or DMEM-with 16 mM glucose (GH). (B) Supernatant sialidase activities in supernatants of the cultures used for panel A. (C) Postinoculation changes in OD600 for 4-h cultures of the wild-type, nanR null mutant, and reversed mutant strains of F4969 grown in control DMEM, DMEM-SL (SL), DMEM-GL (GL), DMEM-SH (SH), or DMEM-GH (GH). (D) Exosialidase activity detection in the supernatants of the cultures used for panel C. (E) qRT-PCR analyses of expression of the nanJ gene (left), the nanI gene (middle), and the nanH gene (right) in control DMEM cultures. All RNA samples were isolated from cells grown for 4 h at 30°C. Transcript levels were determined using 20 ng of RNA. Average CT values were normalized to that of the housekeeping 16S rRNA gene, and the fold differences were calculated using the comparative CT method (2−ΔΔCT). The value of each bar indicates the calculated fold change relative to the control. All panels show mean values for three independent experiments. The error bars indicate standard deviations. *, P < 0.05 compared to the control by ordinary one-way ANOVA.