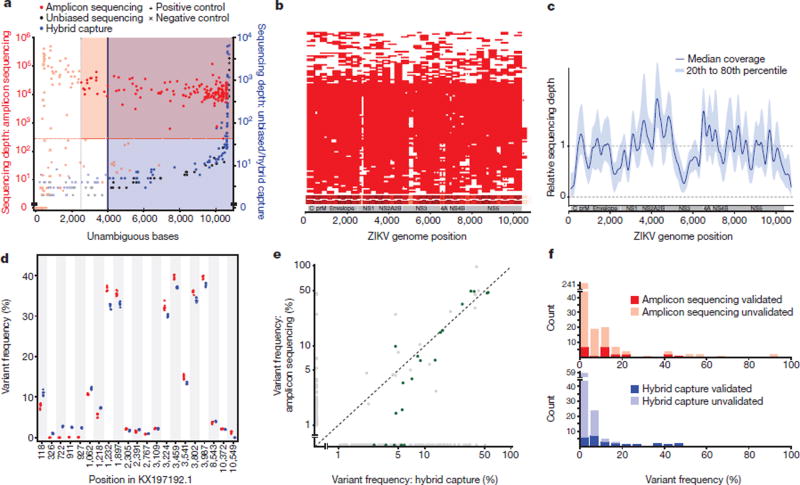
Figure 1. Sequence data from clinical and mosquito samples.
a, Thresholds used to select samples for downstream analysis. Each point is a replicate. Red and blue shading: regions of accepted amplicon sequencing and hybrid capture genome assemblies, respectively. Not shown: hybrid capture positive controls with depth > 10,000×. b, Amplicon sequencing coverage by sample (row) across the ZIKV genome. Red, sequencing depth ≥100×; heatmap (bottom) sums coverage across all samples. White horizontal lines on heatmap, amplicon locations. c, Relative sequencing depth across hybrid capture genomes. d, Withinsample variants for a single cultured isolate (PE243) across seven technical replicates. Each point is a variant in a replicate identified using amplicon sequencing (red) or hybrid capture (blue). Variants are plotted if the pooled frequency across replicates by either method is ≥1%. e, Within-sample variant frequencies across methods. Each point is a variant in a clinical or mosquito sample and points are plotted on a log–log scale. Green points, ‘verified’ variants detected by hybrid capture that pass strand bias and frequency filters. Frequencies <1% are shown at 0%. f, Counts of within-sample variants across two technical replicates for each method. Variants are plotted in the frequency bin corresponding to the higher of the two detected frequencies.