Abstract
Background:
This study aimed to evaluate the role of efflux pump regulator and OXA-23 genes in imipenem resistance Acinetobacter baumannii strains isolated from hospitalized patients in Tehran, Iran.
Methods:
This study was conducted on 60 A. baumannii isolates collected from patients admitted to the Shahid Motahari and Taleghani Hospitals in Tehran during 2013-14. Antibiotic susceptibility tests (AST) and minimal inhibitory concentration (MIC) was determined by broth micro dilution methods according to CLSI 2014 guidelines. The frequency of efflux pump adeRS and OXA-23 genes were detected by PCR and further sequencing.
Results:
The resistance of A. baumannii isolates to tested antibiotics was 100% to cefotaxime, ceftazidime, ceftriaxone, ciprofloxacin, cefepime, piperacillin, meropenem, co-trimoxazole and piperacillin/tazobactam, 97% to imipenem, 94% to gentamicin, 83% to amikacin, 76% to tetracycline, and 0.0% to colistin. The MIC of 58 (96.6%) strains to imipenem was highly decreased in the presence of efflux pump inhibitor (PaβN), by 4 to 64 folds. The adeR and adeS genes were detected in 36 (60%) and 59 (98.3%), respectively and the frequency of OXA-23 gene was 57 (95%) of isolates.
Conclusion:
Existence of adeRS and OXA-23 genes in more than 50% of A. baumannii isolates in this study shows the presumptive role of efflux pump in simultaneous of carbapenemase production. Therefore, using new strategies are required in order to stop the vertical or horizontal exchanges mentioned genes from the resistant A. baumannii isolates to sensitive strains.
Key Words: Acinetobacter baumannii, Antibacterial drug resistance, Efflux pump genes, Carbapenemase, OXA-23
Introduction
Acinetobacter baumannii (A. baumannii) has emerged as a highly problematic hospital-associated pathogen and can be an opportunistic pathogen in humans and affecting people with compromised immune systems (1, 2). This ability is related to their long-term survival in the hospital environment, which increases their opportunities for transmission between patients, either via human reservoirs or via insentient materials. Such infections are often extremely difficult for the clinician to treat because of their widespread resistance to the most common antibiotics (multi drug resistant; MDR) (2). Carbapenems, are the last antibiotics in the most nosocomial infections treatment especially in resistant bacteria (3).
Carbapenem resistance in Acinetobacter spp. may be mediated by one or sum of further mechanisms such as enzyme production (carbapenemase, oxacillinase), loss of outer membrane protein (OMP), alteration of penicillin-binding protein (PBPs) and specific drug efflux pumps (4).
Among them, carbapenemases belongs to types A, B and D of Ambler classification (4). Identification of carbapenemase types in Enterobacteriaceae and Pseudomonas spp. using a phenotypic test to have the common property of hydrolyzing, at least partially, imipenem or meropenem together with other penicillin or cephalosporin antibiotics (4).
Carbapenemases (type D; OXA enzymes) have emerged globally as the main mechanism responsible for this resistance, although metallo-beta-lactamases (type B) are locally prevalent, especially in East Asia. The OXA carbapenemases of Acinetobacter spp. are divided into four phylogenetic subgroups: OXA-23-like; OXA-24-like; OXA-51-like; and OXA-58 (5).
There are five families of efflux pumps, including the multidrug and toxic compound extrusion (MATE) family, the resistance–nodulation–cell division (RND) family, the adenosine triphosphate (ATP)-binding cassette (ABC) family, the major facilitator superfamily (MFS) and the small multidrug resistance (SMR) family, that are associated with multidrug resistance in bacteria. Amongst them, the RND systems are the most prevalent in MDR A. baumannii (6).
The major clinically relevant efflux systems found in A. baumannii, such as AdeABC, AdeIJK and AdeFGH, belong to the (RND) family. AdeABC efflux pump is the first characterized RND system in A. baumannii. Overexpression of the AdeABC and AdeIJK efflux pumps has been shown to pump out aminoglycosides, beta-lactam, fluoroquinolone, tetracycline, macrolide, chloramphenicol, trimethoprim, erythromycin, lincosamides, tigecycline (TGC) and also safranin, pyronine, and sodium dodecyl sulfate (7,8).
The expression of AdeABC efflux pump is tightly regulated by the two-component system, which contains a sensor kinase (SK) AdeS and a response regulator (RR) AdeR, encoded by the adeRS operon transcribed in the opposite direction of AdeABC operon (7).
In this study, we assessed the simultaneous role of the AdeRS genes and OXA-23 carbapenemase among carbapenem resistance A. baumannii isolates from nosocomial infections by the phenotypic and molecular tests.
Materials and Methods
Bacterial isolates
Sixty no repetitive clinical isolates of carbapenem resistant A. baumannii were obtained from 240 clinical samples of two hospitals (Motahari and Taleghani hospitals) in Tehran, Iran during 2013-14.
To ensure the accuracy of genera and species of bacteria, all gram negative nonfermenter bacteria were identified by standard microbial tests(including gram staining, oxidase test, culture on MacConkey agar and TSI (triple sugar Iron agar) and all were confirmed by using Microgen kit (TM Microgen, UK) according to the manufacturer’s protocol.
Antimicrobial susceptibility testing (AST)
Antibiotic susceptibility was tested by disk diffusion on Mueller-Hinton agar (Merck, Germany) according to CLSI 2014 guidelines (all antibiotic disks were purchased from Mast Group Ltd, UK.). E. coli ATCC25922 and K.pneunominia ATCC700603 were evaluated, simultaneously.
Minimum Inhibitory Concentration (MIC)
By broth microdilution method, strains, which were resistant to imipenem during the disk diffusion test, were re-checked and their MIC were determined based on the CLSI 2014 (9). The E. coli ATCC 25922 was used as a quality control strain, simultaneously. Imipenem powder was purchased from Jaberrebne Hayyan co, Iran. Antibiotic preparation was done based on the manufacturer’s recommendations. Antibiotic concentrations were prepared in the range of 256 µg/ml-0.25µg/ml based on CLSI 2014. Bacterial suspension was prepared equal to 0.5 McFarland’s standard turbidity. Antibiotic serial dilution was prepared in a 96-well microtiter plate (Extra Gene-Company-product No.EL-1190-FCS) contained a total volume of 100 µl of each antibiotic dilution and 90 µl of Mueller-Hinton medium with the 10 µl of diluted bacterial inoculums and microplates were incubated at 37°C for 18 to 24 h. The lowest concentration of antibiotic without visible bacterial growth was defined as the MIC (9).
Treatment of the Efflux Pump Inhibitor
As described under microdilution method, changes in susceptibility to imipenem in the presence of 100 µg/ml of the Para amino beta naphtyl amide( PAβN) as an inhibitor was tested. Following the addition of imipenem and the bacterial cell inoculum, 2 µl of the 5-mg/ml stock of either PAβN was added up to 100 µl volume in the microplate . The rest of the procedures were carried out as described above (10). The growth of bacterial suspension in the presence of 100 µl of PAβN was detected for accreditation of the test.
DNA preparation
The DNA was extracted by boiling method (11).
PCR Procedures
The presence of two regulatory (adeS and adeR) genes of the AdeABC system, and one of class D carbapenemases (OXA-23) gene were detected by PCR amplification consequently electrophoresis and further staining by ethidium bromide under UV irradiation. The PCR programs, the sequence of the used primers and the product size were shown in Table 1 and 2, respectively.
Table1.
The PCR programs were used in this study
| Factor | Time | Temperature (ºC) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | AdeS | AdeR | OXA-23 | AdeS | AdeR | OXA-23 | |||||
| Steps | |||||||||||
| Initial denaturation | 5min | 5min | 5min | 94°C | 94ºC | 94°C | |||||
| Denaturation. | 1min | 1min | 25sec | 94ºC | 94ºC | 94ºC | |||||
| Annealing | 1min | 1min | 40sec | 57°C | 57°C | 56°C | |||||
| Extension | 1 min | 1 min | 50sec | 72ºC | 72ºC | 72ºC | |||||
| Final extension | 7min | 7min | 6min | 72°C | 72°C | 72°C | |||||
| Cycles | 30 | 30 | 30 | ||||||||
Table 2.
The primer sequences and the products’ size in this study
| Primer sequences | Genes | Size | Source of references | Company |
|---|---|---|---|---|
|
5′ - GATCGGATTGGAGAACCAGA-3′
5′ -ATTTCTGACCGCATTTCCAT-3′ |
OXA-23-F OXA-23-R |
501 | 13 | Bioneer/Korea |
| 5′-ACTACGATATTGGCGACATT-3 5′-GCGTCAGATTAAGCAAGATT-3′ | AdeR-F AdeR-R |
447 | 12 | Bioneer/Korea |
|
5′-TTGGTTAGCCACTGTTATCT-3′
5′-AGTGGACGTTAGGTCAAGTT-3′ |
AdeS-F AdeS-R |
544 | 12 | Bioneer/Korea |
Sequencing
Finally, all PCR products were sequenced by Bioneer Co, Korea) via Takapuzist company (Tehran, Iran). Sequence analysis was done by Chromas 1.45 software and BLAST in NCBI.
Statistical analysis
This research was a descriptive study. MINITAB16 software was used for the statistical analyses. The P value and confidence intervals were less than 0.05 and 95%, respectively.
Results
Overall, 240 different clinical samples were detected from patients referred to Taleghani and Motahari burn hospitals of Tehran, Iran during 2013-14. Continuously, 60 Acinetobacter spp. confirmed as baumannii were selected randomly. Of included patients, 21 were female and 39 were male. The bacterial isolation based on the hospital wards were 34 were from burn ward, 19 from ICU, 2 of NICU, 2 of nephrology, 1of cardiology, transplant and emergency wards, respectively. In addition, the most frequent isolates were from wound (28), trachea (12), blood (7), sputum (6), stomach and acite fluid (4 each), catheter, CSF, thorax (1), respectively.
Bacterial identification
After Gram staining, oxidase tests, culture on agar MacConkey agar and TSI media and motility. All oxidase-negative, non-fermenter bacteria, non-motile gram-negative bacilli were considered as Acinetobacter spp. Further confirmation was done by microgen kit and the results were read on Color chart based on the manufacturer’s instruction. Based on the software analysis all Acinetobacter spp. were identified as baumannii (Figure1).
Fig. 1.
Schema of microgen kit
Antibiotic profiles of A.baumannii isolates
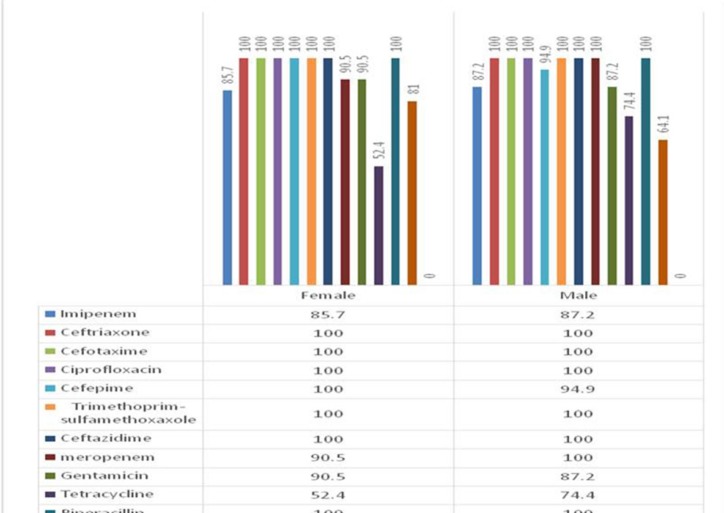
For all 60 A. baumannii isolates, AST was performed and based on the results, 100% of isolates were resistant to ceftriaxone, cefotaxim, ciprofloxacin, trimetoprim-sulfametoxazole, ceftazidim, piperacilin and piperacilin-tazobactam, 96.6% to meropenem and cefepime, 86.6% to imipenem, 88.3% to gentamycin,70% to amikacin and 66.6% to tetracycline. All strains (100%) were sensitive to colistin. All isolates with resistant to more than 3 classes of antibiotics were detected as multi drug resistant (MDR). The frequency of antibiotic resistant based on the gender was shown in the Fig. 2.
Fig. 2.
The frequency of antibiotic resistant based on the gender
MIC of the imipenem and in accompany of PAβN by microdilution method
The MIC of imipenem among A. baumannii resistant isolates are shown in Table 3.
Table 3.
MIC of A. baumannii isolates in this study in comparison to CLSI criteria
| MIC Interpretive Criteria(μg/ml) | |||
|---|---|---|---|
| Species | Resistant | Intermediate | Susceptible |
| Acinetobacter baumannii /CLSI 2014 | ≥16 μg/ml | 8μg/ml | 4 μg/ml ≥ |
| Acinetobacter baumannii/Clinical isolates in this study | 100% | 0% | 0% |
PCR procedure and sequencing
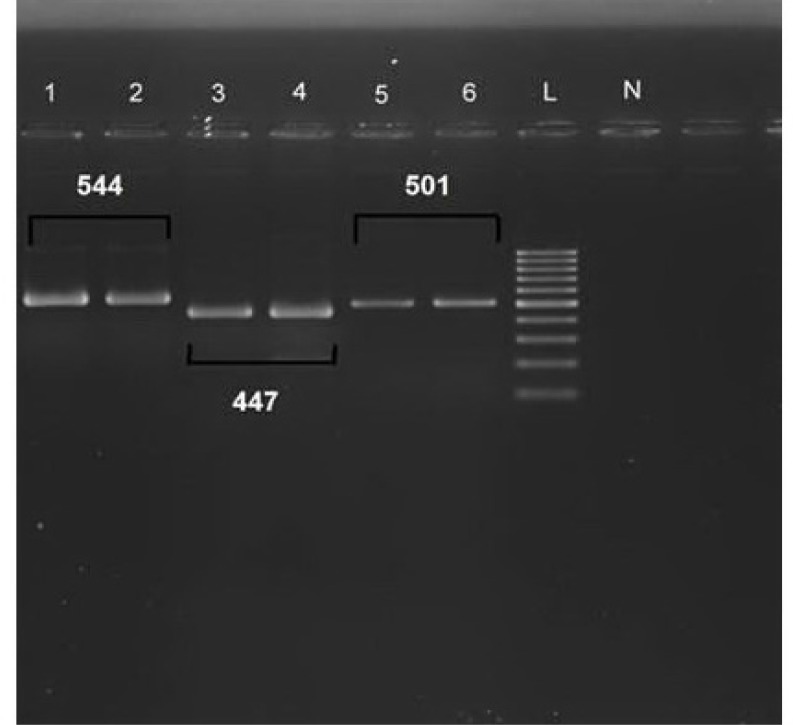
adeR, adeS genes were detected in 36/60 (60%) and 59/60 (98.3%) respectively, also 57/60(95%) of isolates were positive for existence of OXA-23 gene (Figure 3). Later, confirmation of all PCR products was done after comparison to the databank. Gene submission was done, which is in process.
Fig. 3.
The PCR product size of adeS (544bp), adeR (447bp) and OXA-23(501bp) genes in this study. Lanes1, 2: positive (control strain and a clinical sample) for AdeS gene, lanes 3, 4: positive (control strain and a clinical sample) for AdeR gene, lanes 5, 6: positive (control strain and a clinical sample) gene OXA-23.A 100bp plus-Sina Gene ladder was used in lane L for the comparison
Discussion
Existence of 100% resistant to 7 of 14 selected antibiotics by AST showed multidrug resistant among A. baumannii isolates in this study. In addition, by microdilution method 100% of isolates were resistant to imipenem. In addition, in this study the PAβN (an inhibitory compound) effect on the imipenem MIC and decrease it during microdilution method. The effect of efflux inhibitory materials such as PAβN was examined in some other studies too. By adding of 100 µg/mL of PAβN it greatly reduced the MIC of ciprofloxacin from 2 to 8 folds (10). Adding of different concentration of PAβN could reduce the MIC of various antibiotics. Outer membrane protein changes and efflux pump expression together may confer resistance to ertapenem in Enterobacter cloacae. Similarly, the imipenem susceptibility of most isolates was increased in the presence of PaβN, mainly from 4 to 64 folds (12).
The prevalence of AdeS, AdeR, and OXA-23 genes among the A. baumannii isolates of this study was 98.3%, 60% and 95%, respectively.
In China, the relationship between resistance to imipenem and efflux pumps AdeABC, AdeM, AdeDE, were evaluated among the clinical A. baumannii isolates. 80% of A. baumannii strains, which were resistant to imipenem, harbored AdeB, AdeR, Ades and AdeJ genes (13). In accordance to this study, the frequency of AdeRS gene among imipenem resistant A. baumannii strains was investigated and AdeS and AdeR genes were observed in 98.3% and 60% of strains, respectively.
The frequencies of the blaIMP2, blaGES1, blaVIM2, blaOXA-23, blaOXA-51genes were evaluated among A. baumannii strains collected from seven hospitals in Tehran: and the frequency of blaOXA-23 and blaOXA-51 was 94% and 84%, respectively (14). In the present study, although the frequency of genes blaIMP2, blaGES1 and blaVIM2 was not examined, but the frequency of OXA-23 gene was 95%, similarly. This similarity despite the time intervals (about 2 yr difference) between two studies, is another confirmation for the results of the recent study.
The role of RND efflux pumps in A. baumannii resistance to biocides was investigated by RT-PCR (15) and found that increased expression of AdeB and AdeJ genes causes increased resistant to biocides and the frequency of AdeS, AdeR, AdeB genes were observed in 55% of strains (15). In the present study, although gene expression by RT-PCR was not examined, but the frequency of AdeR and AdeS genes were assessed by PCR and they were seen in 60% and 98.3% of strains, respectively. High frequency rate of mentioned genes in this study may be related to increased rate of antibiotic consumption or drug resistant recently.
In Korea, researchers evaluated the role of efflux pump AdeABC and OXA-23 in A. baumannii strains carrying OXA-66 by using PFGE and real time PCR. According to the results, with PFGE technique and digestion by the enzyme SmaI, all isolates had the same restriction pattern. Isolates’ MIC of meropenem were found ≥ 32 mg/L. By real time PCR technique, the expression level of AdeB gene in 6 isolates of A. baumannii was 10 to 40 times greater than the strains susceptible to imipenem and all isolates had blaOXA-23 gene. Finally, they have announced that increased expression of OXA-23 gene and AdeABC efflux pump may have an important role in the emergence of the carbapenems resistance A. baumannii isolates (16).
In accordance to this study, 100% of isolates were resistant to imipenem based on detected MIC by microdilution method and existence of the OXA-23 gene was observed in 95% of isolates. However, the existence of AdeABC was not evaluated in this study but in our other study, existence of AdeAB efflux genes and AdeC gene among A. baumannii isolates were 100% and 85%, respectively (17).
In a Brazilian study, antimicrobial susceptibility was tested by the disk diffusion method and then, samples resistant to imipenem were evaluated by using PCR for the presence of blaOXA-23 gene. Based on the results, 95.4% of the samples had blaOXA-23 gene and the amount of resistance to imipenem and meropenem was 71.4% and 69.7%, respectively (18). Similarly, in this study, sensitivity to antibiotics was tested by disk diffusion method and 86.6% and 96.6% of the strains were resistant to imipenem and meropenem, respectively. In addition, in 95% of strains, OXA-23 gene was found.
In Saudi Arabia, 55 isolates of A. baumannii isolates were collected. These bacteria were resistant to aztereonam, cefepim, ceftazidim and imipenem. Then, E-test for MIC was laid. The presence of ß-lactamase genes were evaluated by PCR. The genes blaOXA-23, blaPER, blaGES, and blaOXA-24 were observed in 60%, 49.1%, 34.5%, and 3.6% of samples.
The genes SHV, CTX-M, VEB, KPC, OXA-58 and metalo-beta-lactamases were not observed. In addition, the samples, which showed high resistance to carbapenems had blaOXA-23 and blaOXA-24 genes (19). In the present study, MIC was determined using microdilution method for imipenem and the samples were undergone PCR to evaluate the presence of genes OXA-23, AdeR, and AdeS. According to CLSI 2014, by using MIC test, resistance to imipenem was shown in 100% of samples. The comparison of two studies can clearly show the direct link of MIC and the presence of OXA-23 gene. On the other hand, the higher of the percentage of OXA-23 existence, shows the higher of the percentages of resistant to imipenem.
In Taiwan, the molecular epidemiology and antimicrobial resistance in Multi Drug Resistance (MDR) Acinetobacter spp were evaluated in 5 hospitals in Taiwan. By using PCR technique, the presence of following genes: intI1, blaIPM, blaVIM, blaOXA-51, blaOXA-23, blaOXA-58, blaOXA-24, blaompC, adeB, adeJ, adeM were evaluated. According to the PCR results, the prevalence of the tested genes was 91% for intI, 57% for blaOXA-23 and 100% for blaompC, adeB, adeJ and adeM. Determining the resistance genes in A. baumannii isolates is necessary to reduce emergence of resistance bacteria (20). In the present study, samples were evaluated by PCR in terms of the presence of the genes OXA-23, AdeR, and AdeS and MIC was determined using microdilution method for imipenem. OXA-23 gene was present in 95% of samples and according to disk diffusion method, 100% of isolates were imipenem resistant. By comparing these two surveys, existence of OXA-23 gene is related to high carbapenem resistance.
In France, RND efflux pumps were evaluated in 14 MDR. A. baumannii isolates and the role of the regulating AdeRS genes were studied in terms of mutation by Multi Locus Sequence Typing (MLST) and Pulsed-Field Gel Electrophoresis (PFGE) methods. Functional mutation in protected areas AdeRS was seen in all the strains in which over expression of AdeABC was present. Mutations were found in two hot spots of AdeS (near histidine 149) and another in the DNA binding domain of AdeR. Also they showed, high incidence of over expressed efflux pump AdeABC among MDR A. baumannii isolates as a result of a variety of individual mutations is in the corresponding of this two-component regulatory system (21). In the present study, although MLST and PFGE techniques were not used, but the existence and the frequency of AdeRS genes were examined by PCR and further sequencing and the presence of AdeRS genes was seen in more than half of the bacterial isolates.
Conclusion
Existence of adeR, adeS (60%, 98.3% respectively) and OXA-23 (95%) genes in more than half of A. baumannii isolates in this study shows the presumptive role of both efflux pump in simultaneous of carbapenemase production during antibiotic resistance of A. baumannii isolates. So, using new strategies are required in order to stop the vertical or horizontal exchanges of the efflux pump and OXA-23 genes from the resistant A. baumannii isolates to sensitive strains. Also by doing AST before any prescription can decrease the emergence of resistant A. baumannii isolates.
Authors’ Contributions
The core idea of this work came from Mojdeh Hakemi- Vala. Dr. Kambiz Davari was another advisor in this project. Helia Ostadasadolah-Malayeri collected and cultured the clinical samples. All the remained phenotypic, molecular tests and manuscript draft was done by her. Final article revision and data analysis was done by Mojdeh Hakemi- Vala.
Acknowledgment
This paper is a part of a student thesis for M.Sc. degree in microbiology presented at Kurdistan Science and Research Branch, Islamic Azad University, Sanandaj, Iran. In addition, we like to say our great thanks to Dr. Ali Hashemi for his assistance in sequencing analysis and gene submission.
Funding/Support
This work was financially supported by microbiology department of Shahid Beheshti University of Medical Sciences, Tehran, Iran and Kurdistan Science and Research Branch, Islamic Azad University, Sanandaj, Iran.
Ethic approval
Doing of this project was accepted in the Ethics Committee of the Kurdistan Science and Research Branch, Islamic Azad University, Sanandaj, Iran.
Conflict of Interest:
The authors declare that there is no Conflict of Interests.
References
- 1.Feng Z, Jia X, Adams MD, Ghosh SK, Bonomo RA, Weinberg A. Epithelial innate immune response to Acinetobacter baumannii challenge. Infect Immun. 2014;82(11):4458–65. doi: 10.1128/IAI.01897-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.E Bergogne-Berezin, K J Towner. Acinetobacter spp as Nosocomial Pathogens: Microbiological, Clinical, and Epidemiological Features. Clin Microbio Rev. 1996;9(2):148–165. doi: 10.1128/cmr.9.2.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Evans BA, Hamouda A, Amyes SG. The rise of carbapenem-resistant Acinetobacter baumannii. Curr Pharm. 2013;19(2):223–38. [PubMed] [Google Scholar]
- 4.Nordmann P, Poirel L. Carbapenem resistance in Acinetobacter baumannii: mechanisms and epidemiology. Clin Microbiol Infect. 2006;12(9):826–36. doi: 10.1111/j.1469-0691.2006.01456.x. [DOI] [PubMed] [Google Scholar]
- 5.Woodford N, Ellington MJ, Coelho JM, Turton JF, Ward ME, Brown S, et al. Multiplex PCR for genes encoding prevalent OXA carbapenemases in Acinetobacter spp. Int J Antimicrob Agents. 2006;27(4):351–353. doi: 10.1016/j.ijantimicag.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 6.Kuo HY, Chang KC, Kuo JW, Yueh HW, Liou ML. Imipenem: a potent inducer of multidrug resistance in Acinetobacter baumannii. Int J Antimicrob Agents. 2012;39(1):33–8. doi: 10.1016/j.ijantimicag.2011.08.016. [DOI] [PubMed] [Google Scholar]
- 7.Sun JR, Perng CL, Chan MC, Morita Y, Lin JC, Su CM, et al. A Truncated AdeS Kinase Protein Generated by ISAba1 Insertion Correlates with Tigecycline Resistance in Acinetobacter baumannii. PLoS One. 2012;7(11):1–14. doi: 10.1371/journal.pone.0049534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Damier-Piolle L, Magnet S, Bremont S, Lambert T, Courvalin P. AdeIJK, a resistance-nodulation-cell division pump effluxing multiple antibiotics in Acinetobacter baumannii. Antimicrob Agents Chemother. 2008;52(2):557–62. doi: 10.1128/AAC.00732-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Performance standards for Antimicrobial Susceptibility Testing (CLSI); Twenty-second Informational Supplement. Twenty-second Informational Supplement. 3. Vol. 32. Wayne, PA, USA: Clinical and Laboratory Standards Institute. CLSI Document M100-S24; 2014. [Google Scholar]
- 10.Valentine SC, Contreras D, Tan S, Real LJ, Chu S, Xu HH. Phenotypic and molecular characterization of Acinetobacter baumannii clinical isolates from nosocomial outbreaks in Los Angeles County, California. J Clin Microbiol. 2008;46(8):2499–507. doi: 10.1128/JCM.00367-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vaneechoutte M, Dijkshoorn L, Tjernberg I, Elaichouni A, Vos PD, Claeys G, et al. Identification of Acinetobacter genomic species by amplified ribosomal DNA restriction analysis. J Clin Microbiol. 1995;33(1):11–15. doi: 10.1128/jcm.33.1.11-15.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Szabo D, Silveira F, Hujer AM, Bonomo RA, Hujer KM, Marsh JW, et al. Outer membrane protein changes and efflux pump expression together may confer resistance to ertapenem in Enterobacter cloacae. Antimicrob Agents Chemother. 2006;50(8):2833–5. doi: 10.1128/AAC.01591-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hou PF, Chen XY, Yan GF, Wang YP, Ying CM. Study of the correlation of imipenem resistance with efflux pumps AdeABC, AdeIJK, AdeDE and AbeM in clinical isolates of Acinetobacter baumannii. Chemother. 2012;58(2):152–8. doi: 10.1159/000335599. [DOI] [PubMed] [Google Scholar]
- 14.Shahcheraghi F, Abbasalipour M, Feizabadi M, Ebrahimipour G, Akbari N. Isolation and genetic characterization of metallo-beta-lactamase and carbapenamase producing strains of Acinetobacter baumannii from patients at Tehran hospitals. Iran J Microbiol. 2011;3(2):68–74. [PMC free article] [PubMed] [Google Scholar]
- 15.Rajamohan G, Srinivasan VB, Gebreyes WA. Novel role of Acinetobacter baumannii RND efflux transporters in mediating decreased susceptibility to biocides. J Antimicrob Chemother. 2010;65(2):228–32. doi: 10.1093/jac/dkp427. [DOI] [PubMed] [Google Scholar]
- 16.Lee Y, Yum JH, Kim CK, Yong D, Jeon EH, Jeong SH, et al. Role of OXA-23 and AdeABC efflux pump for acquiring carbapenem resistance in an Acinetobacter baumannii strain carrying the blaOXA-66 gene. Ann Clin Lab Sci. 2010;40(1):43–8. [PubMed] [Google Scholar]
- 17.Gholami M, Hashemi A, Hakemi-Vala M, Goudarzi H, Hallajzadeh M. Efflux pump inhibitor phenylalanine-arginine ß-naphthylamide effect on the minimum inhibitory concentration of imipenem in Acinetobacter baumannii strains isolated from hospitalized patients in Shahid Motahari burn hospital, Tehran, Iran. Jundishapur J Microbiol. 2015;8(10):1–7. doi: 10.5812/jjm.19048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lais Lisboa Correa, Larissa Alvarenga Batista Botelho, Livia Carvalho Barbosa, Claudio Simoes Mattos, Jupira Miron Carballido, Carmem Lucia Teixeira de Castro, et al. Detection of blaOXA-23 in Acinetobacter spp. isolated from patients of a university hospital. Braz J Infect Dis. 2012;16(6):521–6. doi: 10.1016/j.bjid.2012.10.003. [DOI] [PubMed] [Google Scholar]
- 19.Al-Agamy MH, Shibl AM, Ali MS, Khubnani H, Radwan HH, Livermore DM. Distribution of β-lactamases in carbapenem-non-susceptible Acinetobacter baumannii in Riyadh, Saudi Arabia. J Glob Antimic Resis. 2013;2(1):17–21. doi: 10.1016/j.jgar.2013.08.004. [DOI] [PubMed] [Google Scholar]
- 20.Lin MF, Chang KC, Lan CY, Chou J, Kuo JW, Chang CK, Liou ML. Molecular Epidemiology and Antimicrobial Resistance Determinants of Multidrug-Resistant Acinetobacter baumannii in Five Proximal Hospitals in Taiwan. Jpn J Infect Dis. 2011;64(3):222–7. [PubMed] [Google Scholar]
- 21.Yoon EJ, Courvalin P, Grillot-Courvalin C. RND-type efflux pumps in multidrug resistant clinical isolates of Acinetobacter baumannii: major role of AdeABC overexpression and AdeRS mutations. Antimicrob Agents Chemother. 2013;57(7):2989–95. doi: 10.1128/AAC.02556-12. [DOI] [PMC free article] [PubMed] [Google Scholar]