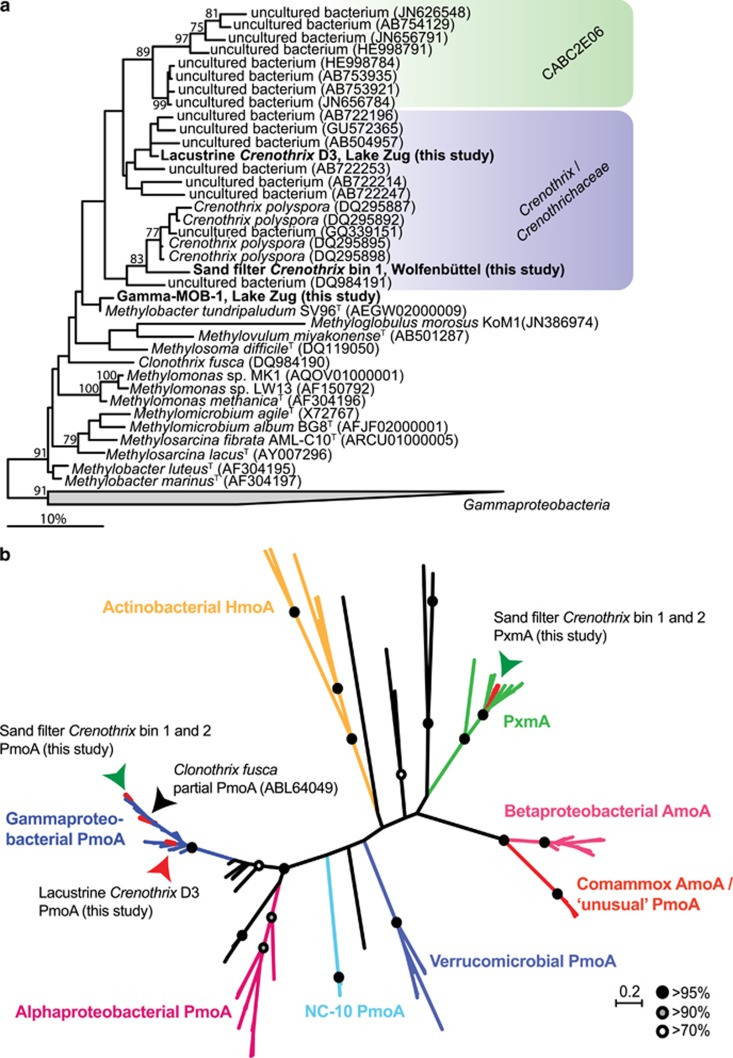
Figure 2.
Phylogenetic tree of Crenothrix 16S rRNA gene and PmoA amino-acid sequences retrieved from Lake Zug and sand filters of the Wolfenbüttel waterworks. (a) Phylogenetic tree of partial 16S rRNA gene sequence retrieved from the lacustrine Crenothrix (909 bp) and from one sand filter Crenothrix (817 bp, bin 1) draft genomes. Note that the 16S rRNA gene sequence of Lake Zug ‘lacustrine’ Crenothrix (but not of the sand filter Crenothrix) is monophyletic with clade CABC2E06. The tree was calculated with the RAxML maximum likelihood program implemented in the ARB package without constraining the alignment by a filter or weighting mask. Bootstrap values >70 (out of 100 resamplings) are shown in front of each node. The taxonomic affiliations indicated by the colored boxes are based on the SILVA SSU reference database (release 123; (Pruesse et al., 2007)). Fourteen type strains spread among gamma-proteobacteria were used as an outgroup. Nucleotide accession numbers are listed in brackets. The bar shows an estimated nucleotide sequence divergence of 10%. (b) Maximum likelihood phylogenetic tree of bacterial PmoA/AmoA amino-acid sequences (135 taxa) showing affiliation of PmoA sequences recovered from the Lake Zug Crenothrix bin (red arrow) as well as of the two sand filter Crenothrix genome bins (green arrows). All three Crenothrix PmoA sequences clustered within the ‘classical’ gamma-proteobacterial PmoA branch. Bootstrap support of total 100 bootstraps are shown in black (>95%), gray (>90%) and white (>70%) circles. Scale bar indicates substitutions per site.