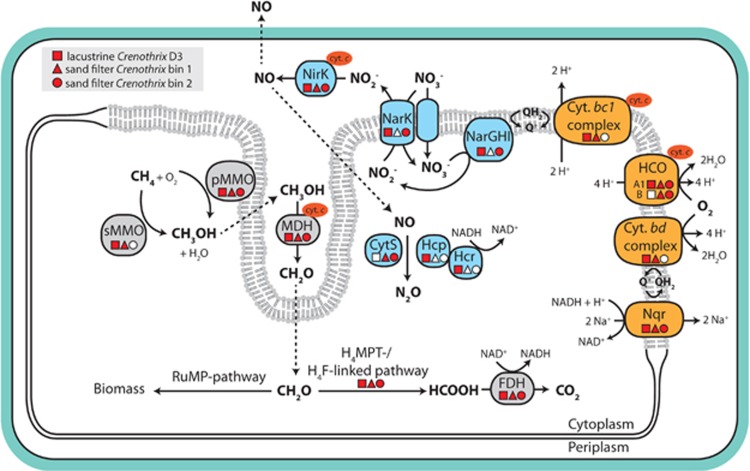
Figure 3.
Genome-inferred metabolic potential of Crenothrix for respiration and methane oxidation. Predicted metabolic potential of the lacustrine Crenothrix as well as of the two sand filter Crenothrix species with respect to its CH4 and N metabolism inferred from the three draft genomes. Indicated are the methane oxidation pathway (gray boxes), the aerobic respiratory chain (orange boxes) and the pathway for nitrate respiration (blue boxes). Genes that were found in the respective Crenothrix genomes (square: lacustrine Crenothrix D3; triangle: sand filter Crenothrix bin 1; circle: sand filter Crenothrix bin 2) are depicted in red, not found in white. Cyt. bc1 complex, cytochrome bc1 complex; Cyt. bd complex, cytochrome bd complex (cydABCD); cyt c., cytochrome c; CytS, cytochrome c’-beta; FDH, formate dehydrogenase; H4F, tetrahydrofolate; H4MPT, tetrahydromethanopterin; HCO, heme copper oxygen reductase (COXI-III); Hcp, hybrid cluster protein; Hcr, NADH-dependent Hcp reductase; MDH, methanol dehydrogenase (xoxF); Nar, nitrate reductase (narGHI); NarK, nitrate/nitrite antiporter (narK); NirS, copper-containing nitrite reductase (nirS); Nqr, sodium-translocating NADH:quinone oxidoreductase; pMMO, particulate methane monooxygenase (pmoCAB); Q, ubiquinone; RuMP, ribulose monophosphate; sMMO, soluble methane monooxygenase (smmoXYBZDC).