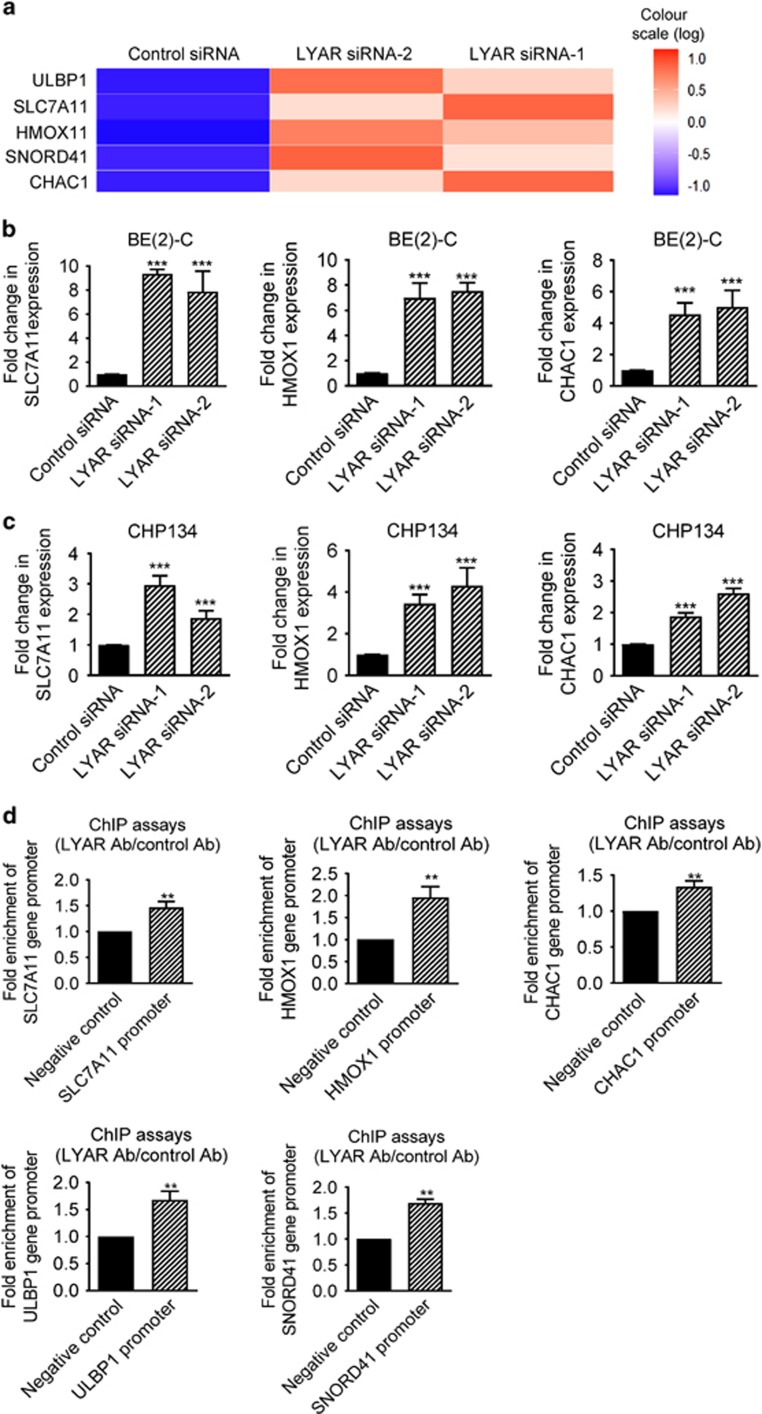
Figure 3.
LYAR regulates oxidative stress gene expression. (a) Genome-wide differential gene expression studies were performed with Affymetrix microarray in BE(2)-C cells 32 h after transfection with control siRNA, LYAR siRNA-1 or LYAR siRNA-2. Heatmap showing the genes up- or downregulated by both LYAR siRNA-1 and LYAR siRNA-2 by more than twofold. Noncoding RNAs and pseudogenes that had never been studied were excluded in the heatmap. (b and c) BE(2)-C (b) and CHP134 (c) cells were transfected with control siRNA, LYAR siRNA-1 or LYAR siRNA-2 for 48 h, followed by RNA extraction. RT-PCR analysis of SLC7A11, HMOX1 and CHAC1 was performed. (d) ChIP assays were performed with a control or anti-LYAR antibody (Ab), followed by PCR with primers targeting a negative control region or the SLC7A11, HMOX1, CHAC1, ULBP1 and SNORD41 gene promoter regions in BE(2)-C cells. Fold enrichment of the LYAR target gene promoters was calculated as the difference in cycle thresholds obtained with the anti-LYAR Ab and with the control Ab. Error bars represented S.E. **P<0.01 and ***P<0.001