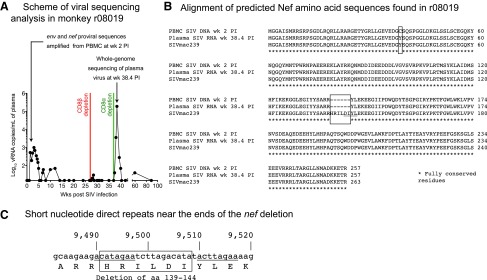
FIG. 6.
Viral sequencing analysis in r08019. (A) Scheme of the viral sequencing analysis performed in r08019. We searched for genetic variation in SIV DNA obtained from cryopreserved PBMC collected at week 2 PI. Viral sequences from nucleotides 7,565–9,979 were amplified, which encode aa 324-879 of Env and the entire Nef protein (aa 1-263). Eleven days after the infusion of the anti-CD8α mAb, when r08019 experienced a VL of 190,000 vRNA copies/ml (week 38.4 PI), we isolated vRNA from plasma and sequenced the SIV genomes circulating at this time point. (B) Alignment of predicted Nef amino acid sequences found in r08019. Conceptual translations of the nef sequences obtained from PBMC-associated SIV DNA at week 2 PI and from plasma vRNA at week 38.4 PI are shown in the upper rows. As a reference, the aa sequence of the SIVmac239 Nef protein is shown in the bottom row. The location of the six-amino acid Nef deletion (139HRILDI144) found in r08019 is indicated by a box. The Y39C substitution present in SIV DNA at week 2 PI is also enclosed by a box. However, this mutation was not detected in the rebounding virus following the CD8α depletion. The sequence corresponding to the SIVmacC8 nef deletion is underlined. (C) Short direct nucleotide repeats near the limits of the deleted region in nef are underlined. These repeated stretches are 8 nucleotides long, seven of which are identical in each repeat. The position of the nucleotides relative to the SIVmac239 genome is indicated on top, and the deleted aa region is enclosed by a box. The nef deletion overlaps with the 3’ long terminal repeat U3 region, which spans nucleotides 9,452–9,978.