Abstract
Membrane proteins (MPs) constitute a third of all proteomes, and contribute to a myriad of cellular functions including intercellular communication, nutrient transport and energy generation. For example, TonB-dependent transporters (TBDTs) in the outer membrane of Gram-negative bacteria play an essential role transporting iron and other nutrients into the bacterial cell. The inherently hydrophobic surfaces of MPs complicates protein expression, purification, and characterization. Thus, dissecting the functional contributions of individual amino acids or structural features through mutagenesis can be a challenging ordeal. Here, we apply a new approach for the expedited protein characterization of the TBDT ShuA from Shigella dysenteriae, and elucidate the protein’s initial steps during heme-uptake. ShuA variants were displayed on the surface of an M13 bacteriophage as fusions to the P8 coat protein. Each ShuA variant was analyzed for its ability to display on the bacteriophage surface, and functionally bind to hemoglobin. This technique streamlines isolation of stable MP variants for rapid characterization of binding to various ligands. Site-directed mutagenesis studies targeting each extracellular loop region of ShuA demonstrate no specific extracellular loop is required for hemoglobin binding. Instead two residues, His420 and His86 mediate this interaction. The results identify a loop susceptible to antibody binding, and also a small molecule motif capable of disrupting ShuA from S. dysenteriae. The approach is generalizable to the dissection of other phage-displayed TBDTs and MPs.
Introduction
Membrane proteins (MPs) perform a wide variety of essential biological functions including intracellular communication, nutrient transport and energy generation. Their intrinsic hydrophobic properties require bicelles, membrane mimics or detergents to solubilize the MP in vitro, and prevent aggregation in an aqueous solution1. This tendency to aggregate complicates structural and biophysical studies of MPs. Therefore, only 1% of the 50,000 unique entries deposited in the Protein Data Bank (PDB) are MPs2. Similarly site-specific mutagenesis as a tool to dissect structure-function relationships of MPs is far less commonly applied compared to soluble proteins3,4.
β -barrel MPs are found on the outer membranes of mitochondria, chloroplasts, and Gram-negative bacteria. Proteins in this class perform perform essential cellular functions including as porins, transporters, enzymes, virulence factors and receptors5. This class of proteins shares common structural features that maintain their stability in the lipid bilayer. Hydrogen bonds connect the 8 to 22 antiparallel β -strands that form the rigid cylindrical barrel. In addition, hydrophobic residues face the lipid bilayer exterior, and polar residues line the interior of the β -barrel. This transmembrane channel can allow passage of polar ligands through the membrane and into the cell6.
For this report, we probed the structure and function relationships of key residues of a protein in a major class of β -barrel membrane proteins, the TonB-dependent transporters (TBDT). This class of MPs is anchored at the bacterial cell surface, and actively transports nutrients through its transmembrane channel into the cell for survival and virulence7. TBDTs use a proton motive force to generate the energy required for transport of essential nutrients across the outer membrane. These transporters directly interact with the TonB protein in the TonB-ExbB-ExbD complex located in the inner membrane to transduce energy from the proton motive force8.
Bacterial pathogens use TBDTs to transport iron across their outer membrane. Iron, an essential nutrient, is utilized for redox oxidation catalysis by a myriad of enzymes. However, the bioavailability of free iron in physiological conditions is severely limited by the insolubility of ferric ions (Fe3+). Additionally, heme-containing proteins sequester free iron in solution. Therefore, bacterial pathogens have evolved specialized iron acquisition systems to fulfill their biological imperative for obtaining iron9.
Iron acquisition systems can be simplified into two general mechanisms. The first mechanism requires a direct contact between the bacterium and iron or iron-containing proteins. In the second mechanism, siderophores and hemophores are secreted into the extracellular medium to scavenge for free iron or heme in the surrounding solution or from the host’s iron/heme-containing proteins. Both iron acquisition systems require a TBDT to transport iron bound siderophores or heme across the bacterial outer membrane10,11.
Shigella dysenteriae, the causative agent of bacillary dysentery, uses a TBDT to acquire heme as its source of iron. The Shigella bacterium infects an estimated 165 million people worldwide by spreading through ingestion of contaminated food or water. About 1 million deaths occur each year from this infection. Victims are often children under the age of 5 and elderly adults12. Antibiotic drug resistant strains are emerging13. Hence, novel therapeutic approaches are needed to combat infections, and reduce its public health burden.
One approach to combat S. dysenteriae infections could target its ability to acquire heme. The TBDT ShuA is necessary for acquiring heme as an iron source through a direct interaction with methemoglobin15,16. This TBDT folds into a typical β -barrel protein with 22 antiparallel β -strands, which are connected by eleven short turns on the periplasmic face and eleven flexible extracellular loops that coalesce at the mouth of the pore opening. The N-terminus forms a globular plug domain that lies within the barrel (Fig. 1). In addition, extracellular loop 7 contains the FRAP (Phe406 – Pro409) and NPNL (Asn434 – Leu437) domains, which are highly conserved in all heme transporters17,18. Although the basic structure of ShuA is known, the initial steps in the heme-uptake mechanism remain largely uncharacterized. Numerous examples of iron-scavenging TBDTs have demonstrated that motifs in its flexible extracellular loops or histidine residues near the pore opening play a role in recognition and binding to the iron source19–21. The structural and functional similarities between ShuA and other TBDTs suggested that ShuA could use a similar mechanism to bind hemoglobin and subsequently extract heme.
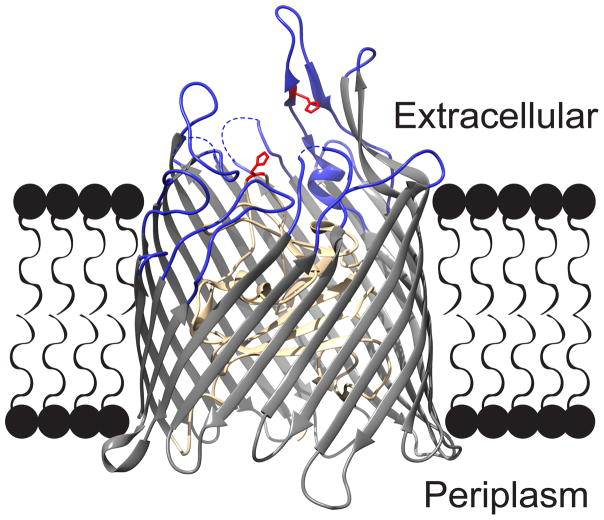
Fig. 1.
The structure of ShuA from Shigella dysenteriae. This model of ShuA protein (PDB 3FHH) highlights the protein’s extracellular loops (blue), histidine residues H86 and H420 (red), and TonB plug domain (tan). The electron density missing in extracellular loops 4, 5, and 10 results from their flexibility (dashes). Molecular graphics were made with UCSF Chimera 14.
To investigate the role of hemoglobin binding by extracellular loops and histidine residues near the pore opening, we introduced site-directed mutations in ShuA’s extracellular loops and key histidines. Previous work in our laboratory has examined the scope of using phage display as a tool for solubilizing different types of membrane-associated proteins, including single or multipass integral MPs and peripheral MPs. These studies have demonstrated that functional membrane proteins can be displayed on the phage surface, particularly the peripheral, monotopic and β -barrel membrane proteins22–25. Here, we apply MP phage display to study structure-function relationships in an MP.
Specifically, site-directed mutagenesis examined the extracellular loops and key histidines proximal to the β -barrel opening of the TBDT ShuA. Each MP variant was displayed on the surface of an M13 bacteriophage, and evaluated for display levels and ligand binding. This phage display approach requires relatively low amounts of protein expression in E. coli for straight-forward purification and rapid analysis of each MP variant. Furthermore the 16.5 mDa bacteriophage can act as a solubilizing handle for ShuA to allow relatively high throughput assays for MP dissection.
Results and Discussion
Expressing the TBDT ShuA on the M13 bacteriophage surface
Previous reports by our laboratory have demonstrated successful display of full-length (70 kDa) ShuA on an M13 filamentous bacteriophage25. For these studies, wild-type ShuA and its site-directed variants were fused through their C-termini to the major coat protein (P8) of the M13 bacteriophage or “phage” through a flexible Gly-Ser linker. A FLAG peptide epitope (amino acid sequence of D Y K D D D D K) was fused to the N-terminus of each ShuA variant to monitor its display levels on the M13 bacteriophage surface. These site-directed mutagenesis studies of a phage-displayed TBDT enable rapid screening of MP variants, and provides the first mutational analysis of hemoglobin binding by ShuA.
After initial failures to provide consistent display of ShuA with each batch of phage, the following conditions were developed. First, the incubation temperature during phage propagation was lowered to 25 °C. At this temperature, the rate of protein synthesis slows, and can allow the β -barrel protein to fold and properly assemble in the inner and outer membranes. Next, adding helper phage to the seed culture upon reaching a cell density of OD600 = 0.4 – 0.5 (not the conventional 0.6) produced more consistent yields of phage and levels of displayed ShuA. Similar approaches are often used to improve the recombinant expression of toxic or membrane-associated proteins27. Additionally, phage-displayed ShuA, wild-type and its variants, were stored in a buffer solution supplemented with N, N-dimethyldodecylamine N-oxide (LDAO) detergent. Fig. SI 1A, Fig. SI 1B, and Fig. SI 1C demonstrate that this zwitterionic detergent is required for display of functional ShuA on the phage surface. LDAO can stabilize native conformational states of MPs by mimicking the lipid bilayer and binding to the hydrophobic regions of the MP28.
Functional characterization of wild-type ShuA on the M13 bacteriophage surface
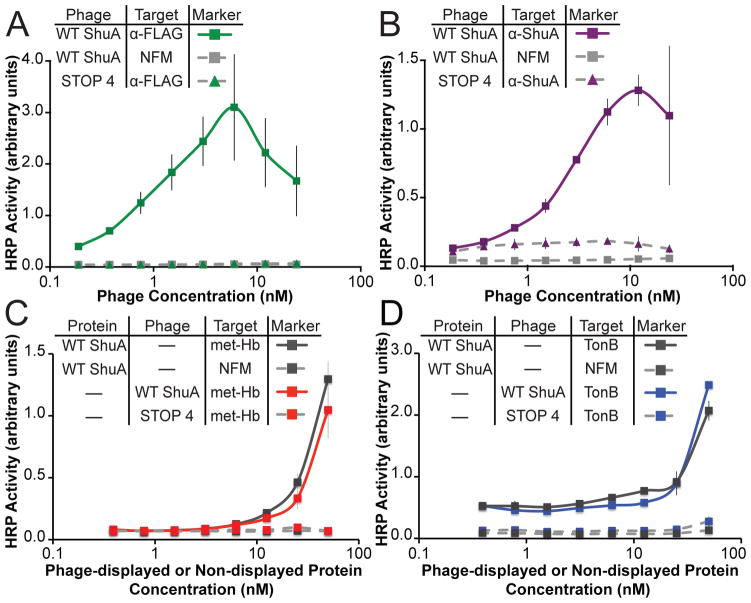
Four phage-based ELISAs were employed to monitor display levels of wild-type ShuA on the surface of M13 bacteriophage and to assess its functional binding. The relative binding affinities of wild-type ShuA to the following immobilized target proteins were assayed: anti-FLAG (α -FLAG) monoclonal antibody, anti-ShuA (α -ShuA) polyclonal antibody, methemoglobin (met-Hb), or TonB. Relative levels of binding to these immobilized targets was quantified through detection with anti-M13 antibody (α -M13) conjugated to horseradish peroxidase (HRP).
To evaluate display levels of wild-type ShuA on the M13 bacteriophage surface, phage-displayed, wild-type ShuA was assayed for binding to immobilized α -FLAG or α -ShuA antibody. Each antibody probes for different regions of wild-type ShuA. The α -FLAG antibody detects the FLAG epitope on the N-terminus. The α -ShuA antibody recognizes a region within the protein coding sequence of phage-displayed wild-type ShuA (Fig. SI 2). Figures 2A and 2B demonstrate that phage-displayed wild-type ShuA binds well to immobilized α -FLAG and α -ShuA antibodies, respectively. As expected, no binding was observed for the two negative controls: non-fat milk used as a blocking agent (NFM) or the bare M13 bacteriophage packaged with an identical phagemid having four stop codons in place of the ORF encoding ShuA (STOP4). These results combined with DNA sequencing of the ShuA phagemid confirm full-length wild-type ShuA successfully displays on the surface of the M13 bacteriophage.
Fig. 2.
Wild-type ShuA displayed on the phage surface and compared to non phage-displayed protein. Phage-displayed wild-type ShuA or STOP4 (negative control phage) was incubated with immobilized A) anti-FLAG antibody (α -FLAG), B) anti-ShuA antibody (α -ShuA), C) methemoglobin (metHb) or D) a 92-residue C-terminal fragment of TonB (TonB). Relative levels of the bound ShuA displayed on phage were quantified by anti-M13 antibody conjugated to HRP. The non phage displayed WT ShuA was detected by an anti-ShuA polyclonal antibody. Each data point represents the average of three replicates, and error bars indicate standard deviation around the mean.
Direct comparison of phage-displayed ShuA with non-phage-displayed ShuA demonstrates the two proteins bind similarly well to two binding partners. The non-phage-displayed ShuA was produced by laborious, conventional methods, and stabilized with detergent using analogous conditions to the phage-displayed protein16. Methemoglobin (metHb) and TonB bind to ShuA on its extracellular and periplasmic sides, respectively16,29. Figures 2C and 2D demonstrate phage-displayed and detergent-solubilized, wild-type ShuA bound to immobilized metHb and TonB with essentially identical apparent binding affinity. Furthermore, treatment of the phage-displayed, wild-type ShuA with 4M urea abolished binding to both binding partners, metHb and TonB, and specifically adsorb to the blocking agent (non-fat milk) or the polystyrene plate, as expected for a denatured protein (Fig. SI 3A and Fig. SI 3B). Taken together, these results demonstrate that ShuA displayed on the M13 bacteriophage is fully folded and functional, including both membrane proximal sides. Having demonstrated both successful display of a membrane protein and sensitive assays for its functions, mutagenesis of ShuA to dissect its mechanism became the next objective.
Site-directed mutagenesis studies of conserved histidine residues of ShuA
Site-directed mutagenesis studies by Wilks and co-workers has identified ShuA His86, in the solvent exposed N-terminal plug domain, and ShuA His420, on extracellular loop L7, extract heme from hemoglobin; the single alanine substitution has greatly reduced activity, and the double Ala-substituted ShuA is nonfunctional16. Given the potential of these highly conserved histidine residues of ShuA to participate in hemoglobin binding and heme transfer, the mutants were studied in our phage display system. The three possible mutants, H86A, H420A, and H86A/H420A, demonstrated high display levels on the viral surface, and were recognized by the ShuA specific antibody. Moreover, all three alanine-substituted ShuA variants have significantly reduced binding to hemoglobin as compared to the wild-type ShuA positive control (Fig. 3). These data demonstrate ShuA H86 and H420 are required for both heme extraction from hemoglobin, and the initial recognition of hemoglobin.
Fig. 3.
Substituting alanine for key histidine residues decreased apparent binding affinity of ShuA for hemoglobin. Phage-based ELISAs for ShuA H86A, H420A, and H86A/H420A were assayed for functional binding to immobilized hemoglobin (A) and display levels by binding to immobilized anti-ShuA antibody (A0 ). The relative binding affinities to hemoglobin for each ShuA variant were calculated as the ratio of A/A0 and normalized to the observed relative binding affinity for wild-type ShuA, the positive phage control. Numbers indicate the percentage loss of hemoglobin binding affinity for each ShuA variant. Error bars were calculated from error propagation for the average of three replicates.
Site-directed mutagenesis studies of the extracellular loops of ShuA
To investigate whether ShuA requires other regions of the protein to interact with hemoglobin, each extracellular loop of ShuA was targeted for two site-directed mutagenesis approaches. First, each extracellular loop was individually deleted. Second, each residue position in the extracellular loops region of ShuA was substituted for alanine. The phage-displayed variants were then examined for display levels and the ability to bind to hemoglobin. The two mutagenesis approaches ensure that the observed phenotype results from loss of critical residues required for the interaction, and not structural perturbation. For example, removal of a critical motif in the extracellular loop region can be expected to abrogate binding to hemoglobin. In theory, the two approaches should provide complementary and identical results.
The crystal structure of apo-ShuA (PDB 3FHH)18 was used to design the deletion and alanine-substituted variants (Table 1). Each construct was generated using overlap extension PCR. At a long 32 residues in length, extracellular Loop 7 was split in half, and two deletion and two alanine substitution variants were designed to minimize structural perturbation. The two halves were designated loop 7A (residues 408 – 422) and loop 7B (residues 423 – 439). For each variant, three to six loop residues remained unchanged. These unchanged residues can ensure that the targeted loop still form a β -turn, which typically requires a minimum of three residues30. DNA sequencing verified successful mutagenesis. In this report, the extracellular loop deletion and alanine-substituted phage-displayed variants will be referred to as ’phage-displayed ΔL1’ or ’phage-displayed Ala-L1’, respectively; the number indicates the mutated extracellular loop, Δ indicates deletion of the targeted extracellular loop, and Ala indicates substitution of the targeted loop residues with alanine, respectively.
Table 1.
Design of extracellular loop deletion and alanine substitution ShuA variants.
| Extracellular Loop | Residue Positions | Amino Acid Sequence* |
|---|---|---|
| 1 | 146 – 151 | G T G D H S |
| 2 | 176 – 189 | R G D L R Q S N G E T A P N |
| 3 | 225 – 238 | K N P Q T V E A S E S S N P |
| 4 | 279 – 286 | Q N T G S S G E |
| 5 | 235 – 226 | P G G A T T G F P Q |
| 6 | 376 – 372 | D G Y K D |
| 7A | 408 – 422 | A P T M G E M Y N D S K H F S |
| 7B | 423 – 439 | I G R F Y T N Y W V P N P N L R P |
| 8 | 483 – 487 | D F A A A |
| 9 | 526 – 537 | D T D T G E Y I S S I N |
| 10 | 564 – 578 | D R S T H I S S S Y S K Q P G |
| 11 | 604 – 623 | G N A F D K E Y W S P Q G I P Q D G R N |
Bold residues were targeted for mutagenesis either as a deletion or alanine substitution
Small variations in the composition of the loop deletion mutants were explored to examine the sensitivity of MP display levels to changes involving just a couple of residues. A bottleneck in conventional MP mutagenesis studies is empirically identifying the optimal length for a loop deletion variant without perturbing its expression levels or tertiary structure. ShuA loop deletion variants, ΔL6, ΔL7 and ΔL11 exhibited display levels similar to wild-type ShuA (Fig. SI. 4A). These extracellular loop deletion variants were targeted for further mutagenesis by deleting an additional 1 – 2 residues, and the display levels were evaluated. These second versions of ShuA ΔL6, ΔL7 and ΔL11 loop deletion variants could not be displayed on the phage surface (Fig. SI 4B). We hypothesize that the more drastic deletions interfere with protein folding and prevent ShuA variant display. By establishing a method that rapidly evaluates varying lengths of ShuA extracellular loop deletion variants, the bottleneck in experimentally probing MP structure is shifted to mutagenesis, not protein expression or purification. The twelve extracellular loop deletion variants chosen for this report, ShuA ΔL1 through ΔL11, were propagated in identical conditions and assayed in parallel to allow direct comparisons. The phage-displayed, wild-type ShuA provided a positive control and the 12 variants bound to the immobilized α -FLAG antibody at similar levels (Fig. SI 5A and Fig. SI 5B); the modest degree of variablilty was accounted for by normalization as described previously31. The results confirm all loop variants used for this study are displayed on the bacteriophage with similar levels.
Having established similar levels of phage-displayed ShuA ΔL1 through ΔL11 variants were further evaluated for display levels with an α -ShuA antibody ELISA. With the exception of ShuAΔL7A and ΔL7B, each of the extracellular loop deletion variants bound to the α -ShuA antibody at similar levels as phage-displayed wild-type ShuA (Fig. SI 5C, Fig. SI 5D and Fig. SI 5E). Deletion of loops 7A and 7B decreased apparent binding affinity to the α -ShuA antibody. Notably, display levels for ShuA ΔL7A and ΔL7B variants remain about the same, as verified by α -FLAG ELISA. Furthermore, the two loop deletion ShuA variants maintained protein function, as demonstrated through binding hemoglobin. The results suggest that deleting regions of Loop 7 interferes with an α -ShuA antibody epitope.
Each extracellular loop deletion variant was then tested for hemoglobin binding. Site-directed mutagenesis studies of TBDT HgbA from Haemophilus ducreyi demonstrate that motifs in the extracellular loops are required to bind hemoglobin, a prerequisite to heme transfer19,32. Thus, removal of an essential extra-cellular loop region of ShuA was expected to disrupt the ShuA-hemoglobin interaction. However, wild-type ShuA and its loop deletions ΔL1 through ΔL11 bind hemoglobin with similar binding affinity (Fig. SI 5F, Fig. SI 5G and Fig. SI 5H). Thus, unlike other TBDTs, ShuA does not rely on a specific extracellular loop to recognize and bind to its ligand, hemoglobin.
To control for the possibility of severe structural perturbations caused by loop truncation, these experiments were repeated with less drastic modifications to ShuA. All residues formerly targeted for deletion were substituted with alanine. As before, the 12 alanine-substituted ShuA variants were displayed on the bacteriophage surface and assayed for display levels and function. The results confirm no specific extracellular loop is required for the ShuA-hemoglobin interaction (Fig. 4A). Furthermore, both alanine-substituted ShuA L7 variants, ShuA Ala-7A and Ala-7B, exhibited similar apparent binding affinity as wild-type ShuA to the immobilized α -FLAG and methemoglobin targets. Thus, display and function for the Ala-substituted loop variants are not perturbed. The phage-displayed ShuA Ala-L7A and Ala-L7B bound the α -ShuA antibody with decreased apparent affinity as wild-type ShuA. The loss of α -ShuA antibody recognition to the four ShuA L7 variants, two deleted and alanine substituted L7 halves, stems from ablation of an antibody binding epitope present within Loop 7 of ShuA (Fig. SI 2). The antibody target mapping demonstrated here suggests that vaccine development could focus on Loop 7.
Fig. 4.
Binding to hemoglobin by ShuA variants with modified extracellular loops. Phage-based ELISAs for ShuA extracellular loop variants were assayed for functional binding to immobilized hemoglobin (A) and binding to immobilized anti-ShuA antibody (A0 ). The relative binding affinities to hemoglobin for each ShuA variant were calculated as the ratio of A/A0 and normalized to the observed relative binding affinity for the positive phage control, wild-type ShuA (WT). A) Mutations to ShuA extracellular loops, either deletion (gray) or alanine substitutions (white) targeted the indicated individual loops. These mutations resulted in no significant change in apparent binding affinity for hemoglobin. B) ShuA variants with alanine substitutions targeting multiple extracellular loops (Ala-L3/Ala-L5, Ala-L3/Ala-L6, Ala-L5/Ala-L6 and Ala-L3/Ala-L5/Ala-L6) demonstrate a 30% decrease in apparent relative binding affinity to hemoglobin. Error bars were calculated from the error propagation equation with three replicates.
We next turned our attention to additive effects from multiple loops contributing to hemoglobin binding. To identify if multiple extracellular loops of ShuA are required for hemoglobin binding, 2 or 3 loops of ShuA were substituted simultaneously with alanine. For these studies, extracellular loop variants ShuA Ala-L3/Ala-L5, ShuA Ala-L3/Ala-L6, ShuA Ala-L5/ Ala-L6, and ShuA Ala-L3/ Ala-L5/ Ala-L6 were displayed on the bacteriophage surface and assayed for display levels and function, as described above. The results demonstrate extracellular loops may be required for binding hemoglobin by cumulatively weak interactions, without the need for a specific recognition motif (Fig. 4B). Thus, unlike other TBDTs, ShuA does not rely on a single extracellular loop to recognize and bind to hemoglobin, but each extracellular loop cumulatively contributes a small amount of binding energy to interact with hemoglobin.
Protein-Protein Interaction Studies with Purified ShuA protein variants and Hemoglobin
To verify observations made with phage-displayed protein accurately represents the protein removed from the phage surface, ShuA protein variants H86A/H420A, Ala-L11, and wild-type were recombinantly overexpressed in the outer membranes of E. coli bacteria. The MPs were isolated by ultracentrifugation, solubilized in LDAO detergent micelles, and purified by multi-column FPLC before visualization by SDS-PAGE analysis. Analytical SEC of the purified protein demonstrates the effective removal of any aggregated protein for subsequent analysis (Fig. 5A). A single band with a 73 kDa molecular mass and >95% homogeneity was observed for the three overexpressed proteins, including wild-type ShuA (Fig. 5A). The three ShuA variants were further examined for overall secondary structure by circular dichroism (CD). Wild-type ShuA served as a positive control. Analysis of the CD spectra of ShuA H86A/H420A and ShuA Ala-L11 demonstrate that secondary structure was not perturbed by the introduced mutations (Fig. 5B). Next, the three purified proteins were tested for binding to hemoglobin; wild-type ShuA serves as the positive control and NFM as the negative control. As observed for phage-displayed proteins, the ShuA Ala-L11 variant bound to hemoglobin with similar binding affinity as wild-type ShuA (Fig. 5C). The measured EC50 values for ShuA Ala-L11 and wild-type binding to hemoglobin was ≈0.53 μ M as estimated using non-linear regression analysis of the dose response curve. This assay confirms the results with phage-displayed ShuA that extracellular loops can contribute to hemoglobin binding by having cumulative weak interactions. In addition, ShuA H86A/H420A bound to hemoglobin significantly less compared to wild-type ShuA with an EC50 value of ≈6.9 μ M. Again, these results support the phage-based studies reported here showing that the conserved histidine residues play an essential role in hemoglobin binding and subsequent heme extraction.
Fig. 5.
Non-phage-displayed ShuA protein variants binding to hemoglobin. A) Representative size exclusion chromatogram of a ShuA variant. Inset: Coomassie Blue stained 12% SDS-PAGE gel of purified ShuA variants. Lane 1, molecular weight markers; Lanes 2–4, purified wild-type ShuA, Ala-L11 and H86A/H420A, respectively. B) Circular dichroism spectra of purified ShuA variants. Spectra of ShuA wild-type, ShuA L11-Ala and ShuA H86A/H420A were recorded at 25 °C in 50 mM potassium phosphate (pH 7.5) and 50 mM NaCl containing 0.05% (w/v) LDAO at a final protein concentration of 5 μ M. C) ELISA assay measuring hemoglobin binding by purified ShuA variants. Purified ShuA protein variants wild-type, L11Ala, and H86A/H420A were applied to immobilized methemoglobin-coated wells on a microtiter plate at the indicated concentrations. The amount of ShuA bound to immobilized hemoglobin was detected by through binding by anti-ShuA polyclonal antibody and secondary anti-rabbit HRP-conjugated antibody, and measured at A450 . Error bars were calculated as the standard deviation of three replicates.
To chemically probe the role histidine residues play in hemoglobin binding, wild-type ShuA was treated with a histidine-specific reagent, diethylpyrocarbonate (DEPC). Binding to hemoglobin for the treated ShuA was compared to an untreated, otherwise identical sample. Wild-type ShuA lost about 60% of its activity for binding to hemoglobin after treatment with 200 μ M DEPC (Fig. 6). This observed loss of activity is similar to the mutagenesis studies with phage-displayed and recombinantly expressed ShuA H86A/H420A. To determine if the DEPC modification of the histidine residue(s) could be reversed, a DEPC-treated sample was further treated with hydroxylamine, a reagent widely used to deacylate N-carbethoxyhistidines33. As shown in Figure 6, DEPC-treated wild-type ShuA loss 60% hemoglobin binding activity, and its activity could be partially restored to a 40% loss through further treatment with hydroxylamine. The failure of hydroxylamine to restore hemoglobin binding activity of the DEPC-modified wild-type ShuA likely is the result of formation of N,N-dicarbethoxyhistidine. This double-substituted adduct of His is irreversibly formed in excess DEPC33. Although histidines residues are clearly important for hemoglobin binding as shown by mutagenesis and DEPC treatment, further studies could examine off-target effects by the promiscuous DEPC on tyrosine and other residues.
Fig. 6.
DEPC treatment of wild-type ShuA protein abolishes hemoglobin binding. Three samples of wild-type ShuA protein (5μ M) in 50 mM potassium phosphate buffer, 50 mM NaCl (pH 7.0) were treated with and without 200 mM DEPC. The positive control was an untreated sample. Following treatment one of the DEPC-treated samples was subsequently treated with 100 mM hydroxylamine. The three samples were assayed in a hemoglobin ELISA as described above. The relative binding affinity is normalized to the positive control. Error bars indicate the standard deviation around the mean (n=3).
Conclusion
In summary, we sought to identify specific surface-exposed extracellular loops of ShuA that are important for hemoglobin binding. For these studies, 24 variants of ShuA carrying deletion or alanine substitutions in each of the surface-exposed extracellular loops were displayed on an M13 bacteriophage surface and evaluated for display levels and hemoglobin binding. We have shown that individual loops do not significantly contribute to hemoglobin binding. Instead, ShuA may be binding to hemoglobin by cumulative weak interactions between its extracellular loops, and two highly conserved histidine residues, H86 in the plug domain and H420 in extracellular loop L7. The amino acid composition of the extracellular loops of ShuA consists of a mixture of hydrophobic, acidic and basic residues that vary in length from 6–32 residues. Short regions of the extracellular loops of ShuA can interact with hemoglobin via cumulatively weak hydrogen bonds, electrostatic, and hydrophobic interactions. The transient interaction between the extracellular loops of ShuA and hemoglobin may help orient hemoglobin at the pore opening for effective heme extraction by the conserved histidine residues.
This was demonstrated through phage-displayed mutagenesis studies that exhibited a 30% reduction in hemoglobin binding activity when 3 out of the 11 extracellular loops were alanine-substituted. Hemoglobin ELISAs studies with phage-displayed and non-displayed recombinantly expressed ShuA H86A/H420A that exhibited a significant reduction to hemoglobin binding affinity. In addition, to a 60% reduction in hemoglobin binding activity with wild-type ShuA treated with the histidine-specific modifier, DEPC, and a partially restoration in hemoglobin binding activity upon hydroxylamine treatment of DEPC inhibited wild-type ShuA.
Histidines are common axial ligands of heme-iron in proteins34,35. TBDTs in the heme-uptake pathway of several Gram-negative bacteria from Yersinia enterocolitica (HemR), Yersinia pestis (HmuR), E. coli (ChuA), and Poryphyromonas gingivalis (HmuR) demonstrate that histidines are essential for heme binding, extraction and utilization17,36. For these structurally similar receptors, it is apparent that the combination of conserved FRAP/NPNL domains and histidine, or distinct binding sites enable them to either functionally recognize specific heme-protein complexes, such as Hb or Hb-haptoglobin, or recognize multiple heme-protein complexes with a common motif. The point mutations (H86A and H420A) and double mutations (H86A/H420A) illustrate the key role played by these positions in the initial hemoglobin-binding step for heme-uptake. These residues are then required for subsequent heme coordination, and transfer16. These results are in alignment with studies of Y. enterocolitica (HemR) where two histidine residues (H128 and H461) are essential for heme utilization by the heme receptor HemR; furthermore, P. gingivalis uses histidines (H95, H434) in HmuR for a similar role17,36.
Additional structural characterization studies are required to elucidate the mechanism histidines play in forming the ShuA-hemoglobin complex and extracting heme from hemoglobin. Although specific motifs in the extracellular loop regions did not contribute significantly to hemoglobin recognition and binding, they may play an essential role in heme transport through the β-barrel and into the periplasmic space.
This report demonstrates the potential of phage-based mutagenesis for protein engineering and structure-function studies of membrane-associated proteins. As demonstrated here, MP phage display opens the TBDT class of β -barrel proteins to detailed dissection. TBDT represent excellent targets for therapeutic and vaccine development, as they necessarily remain exposed and available for targeting. However, methods to identify key features for such proteins have lagged behind developments in high throughput drug discovery. The phage-based mutagenesis method reported can be generalizable to many proteins in this class. Furthermore, combining phage display with drug discovery could accelerate and allow more detailed elucidation of structure-activity relationships.
Materials And Methods
Cloning the ShuA variants for phage display and protein expression
Each extracellular loop deletion or alanine-substituted variant of ShuA was constructed by overlap extension PCR with plasmid pEShuA16 as the template. The resulting PCR product, with flanking 5’ NsiI and 3’ NcoI restriction sites, was sub-cloned between the secretion signal peptide and the gene encoding the major coat protein (g8p) for phage display studies, as previously reported25. The phagemid vector encoding the extracellular loop deletion or alanine-substituted variants of ShuA served as the template for subsequent PCR amplification with flanking 5’ MscI and 3’ XhoI restriction sites. The resultant PCR product was then sub-cloned into the pET22b (Novagen) plasmid vector in-frame with a sequence encoding an N-terminal His6 Tag for recombinant protein expression and purification. Genewiz LLC performed the DNA sequencing. Sequences of the oligonucleotides used for mutagenesis appear in the Supporting Information (Table 1).
Purification of the phage-displayed ShuA variants
The phage-displayed ShuA variants were purified as described previously25 with the following changes. The E. coli SS320 cell strain (Lucigen) was transformed with the pM1165a phagemid encoding the extracellular loop deletion or alanine-substituted variants of TBDT ShuA. The seed culture was grown to an OD600 = 0.4–0.5 and infected with M13 KO7 helper phage (GE Healthcare) to package the viral particles. The seed culture was transferred to 250 mL 2YT media supplemented with 40 μ g /mL kanamycin and 25 μ g /mL carbenicillin and incubated at 30 °C. After the second phage precipitation step, the phage-displayed ShuA variants were resuspended in ShuA buffer (50 mM Tris, pH 7.8, 50 mM NaCl, and 0.04 to 0.05% (w/v) LDAO. Phage concentrations were estimated spectrophotometrically (1 OD268 = 8.31 nM).
Phage-based ELISAs
Phage-based ELISAs were performed as described previously22 with the following changes. To assess display and functionality of the phage-displayed ShuA variants, specific wells of a 96-well Nunc maxisorb plate were coated for 14–16 h at 4 °C with 100μ L/ well of PBS solution containing one of the following: monoclonal anti-FLAG M2 antibody (1:1000 dilution) (Agilent Technologies), a polyclonal anti-ShuA antibody from rabbit serum16 (1:2000 dilution), methemoglobin purified from red blood cells (10 μ g/ml), as described previously37, or His6 -tagged TonB protein (10 μ g/ml). The blocking agent was 0.2% w/v non-fat milk (NFM).
Non-phage-displayed ELISA
Non-phage-displayed ELISAs were performed as described previously22, with the following changes. The target protein methemoglobin from red blood cells (10 μ g/ml) was coated on a microtiter plate. After blocking with 0.2% w/v NFM in PBS, the indicated concentrations of ShuA variants (WT, H420A/H86A or Ala-L11) were added to the wells. An anti-rabbit conjugated to horseradish peroxidase secondary antibody (1:2000 dilution) (Thermo Fisher Sci) was used to measure ShuA-hemoglobin binding. A sigmoidal dose response curve was obtained to calculate EC50 values for purified ShuA variants binding to immobilized met-hemoglobin. Non-linear regression analysis was performed with GraphPad Prism.
Cloning of TonB protein
A 92-residue C-terminal fragment of the tonB gene (encoding residues 142 – 239) from S. dysenteriae was PCR-amplified from the expression plasmid pND3416 and cloned into the expression vector pET28a with an N-terminal His6 Tag and flanking 5’ NdeI and 3’ XhoI sites.
Purification of TonB protein
BL21 (DE3) cells were transformed with pET28a-TonB expression plasmid and incubated for 12 h at 37 °C. An overnight seed culture was transferred to 1 L Terrific Broth (TB) media supplemented with kanamycin (40 μ g/ml) and incubated at 37 °C with shaking until the cells reached an OD600 = 0.6. The cells were induced by addition of 1 mM IPTG, and incubated at 25 °C with shaking for 12 h. The cells were harvested by centrifugation (6,000 g, 15 min) resuspended in lysis buffer (50 mM Tris pH 7.7, 300 mM NaCl, 10 mM imidazole) supplemented with 1X halt protease inhibitor (ThermoFisher Scientific), 1 mM phenylmethylsulfonyl fluoride and 10 mM benzamidine before lysis by sonication. The cellular debris was removed by centrifugation (30,000 g, 45 mins). The supernatant containing solubilized TonB protein was applied to a nickel-bound IMAC resin (Bio-Rad) equilibrated with lysis buffer, washed with 10 column volumes of wash buffer (50 mM Tris pH 7.7, 300 mM NaCl, 50 mM imidazole), and eluted with 10 column volumes of wash buffer supplemented with 250 mM imidazole. Purified TonB protein was further purified by size exclusion chromatrography and analyzed for purity by SDS-PAGE (SI Fig. 4), which demonstrated >95% homogeneity.
Expression and purification of ShuA protein variants
Seed cultures of E. coli C41 (DE3) cells transformed with pET22b plasmid DNA encoding ShuA WT, ShuA H86A/H420A or ShuA Ala-L11 were inoculated in 2YT medium supplemented with 50 μ g/mL carbencillin for 14–16 h at 30 °C. The seed cultures were transferred to TB media supplemented with 50 μ g/mL carbenicillin, and incubated at 30 °C until the cell density reached OD600 = 0.5. Protein expression was induced by the addition of 1 mM IPTG at 30 °C for 22 h. Cells were harvested by centrifugation (6,000 g, 15 min), resuspended in buffer A (50 mM Tris-HCl, pH 7.8, 50 mM NaCl, 12% glycerol) supplemented with 1X Halt protease inhibitor (Thermo Fisher Scientific), DNaseI (20 μ g/mL) before lysis by lysozyme treatment and sonication. The cell debris was removed by centrifugation (20,000 g, 15 min). The total cellular membranes were pelleted by ultracentrifugation (105,000 g, 1 h). The inner membrane proteins were solubilized in buffer A supplemented with 1% (v/v) Triton X-100 and 1% (w/v) N-lauroylsarcosine, and stirred at 4 °C for 3 h. The outer membrane was pelleted by centrifugation (105,000 g, 1 h), and solubilized in Buffer A containing 1X Halt protease inhibitor and 1% (w/v) LDAO, and stirred at 4 °C for 16 h. The cellular membrane debris was pelleted by centrifugation (30,000 g for 1 h), and the clarified supernatant was applied to nickel-bound IMAC resin (Bio-Rad) equilibrated with Buffer B (50 mM Tris-HCl, pH 8.0, 50 mM NaCl, 0.04 to 0.05%(w/v) LDAO, 10 mM imidazole, and 12% glycerol). The column was washed with 10 column volumes of wash buffer (50 mM Tris-HCl, pH 7.8, 50 mM NaCl, 0.04 to 0.05% (w/v) LDAO, 20 mM imidazole, and 12% glycerol), and eluted with 10 column volumes of wash buffer supplemented with 125 mM imidazole. A fraction of the purified ShuA protein was analyzed by SDS-PAGE, which demonstrated >95% homogeneity after purification by Ni-IMAC. The remaining protein was dialyzed into Buffer A supplemented with 0.04 to 0.05% (w/v) LDAO and concentrated to 0.5–1 mL with a 50 MWCO filtered micro concentrator (EMD millipore). The concentrated ShuA was purified from protein aggregates with a Superdex 200 10/300 GL column equilibrated in 50 mM Tris-HCl (pH 7.8) containing 150 mM NaCl. Purified fractions were analyzed by SDS-PAGE and pooled for subsequent experimental analysis.
Circular dichroism of ShuA variants
Following overexpression and purification, the His6 -tagged wild-type ShuA and variants, Ala-L11 and H86A/H420A, were dialyzed into 50 mM NaH2 PO4, pH 7.8, 50 mM NaCl, and 0.04 to 0.05%(w/v) LDAO. A circular dichroism spectrum from 190 to 260 nm was acquired for each protein sample (5 μ M) on a Jasco spectropolarimeter (model J810) with a 0.1 cm path-length cell, 0.2 mm resolution and 1.0 cm bandwidth at 23 °C. A total of 20 consecutive scans were accumulated for analysis. DichroWeb was used to analyze the data38.
Determining relative binding affinities for phage-displayed ShuA variants
A single 96-well microtiter plate was used to simultaneously measure display levels and hemoglobin binding levels for various ShuA variants. The apparent binding affinity was calculated as the ratio between the binding levels to immobilized hemoglobin for evaluating function (A) and the binding levels to immobilized anti-ShuA antibody for monitoring the display levels on the phage surface (A0 ), as described previously 31. The apparent relative binding affinities were determined for each ShuA variant, and normalized to the apparent binding affinity of wild-type ShuA, which served as the positive control.
Inactivation of wild-type ShuA protein by diethyl pyrocarbonate (DEPC)
A stock solution of 6.9 M DEPC was freshly prepared by diluting DEPC with cold absolute ethanol (1:19, v/v). A solution of 0.5 mM DEPC in 0.1 M sodium phosphate buffer (pH 6.5) was freshly prepared before use and kept on ice. The reaction mixture was prepared with a final concentration of 5 μ M wild-type ShuA protein and 0.2 mM DEPC, and incubated at 25 °C for 10 mins.
Hydroxylamine treatment of inactivated wild-type ShuA
After 10 min incubation of wild-type ShuA with 200 mM DEPC, 0.5 M hydroxylamine was added to a final concentration of 0.1 M. The mixtures were then incubated at 4 °C for 40 min, and analyzed by a protein ELISA to measure hemoglobin binding.
Supplementary Material
Acknowledgments
The authors are grateful for financial support from the National Institutes of Health (Grant GM100700-01 to G.A.W. and grant GM057353 to T.L.P.). LM received the Miguel Velez Scholarship at UCI. We also gratefully acknowledge Dr. Angela Wilks for helpful discussions and generously providing polyclonal rabbit anti-ShuA antibodies, and ShuA protein expression constructs, wild-type pEShuA and H86A/H420A pEShuA.
Footnotes
Electronic Supplementary Information (ESI) available: [details of any supplementary information available should be included here]. See DOI: 10.1039/b000000x/
References
- 1.Seddon AM, Curnow P, Booth PJ. Biochim Biophys Acta. 2004;1666:105–117. doi: 10.1016/j.bbamem.2004.04.011. [DOI] [PubMed] [Google Scholar]
- 2.White SH. Nature. 2009;459:344–346. doi: 10.1038/nature08142. [DOI] [PubMed] [Google Scholar]
- 3.Zhang HT, Unal H, Desnoyer R, Han GW, Patel N, Katritch V, Karnik SS, Cherezov V, Stevens RC. J Biol Chem. 2015;290:29127–29139. doi: 10.1074/jbc.M115.689000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Popov M, Tam LY, Li J, Reithmeier RA. J Biol Chem. 1997;272:18325–18332. doi: 10.1074/jbc.272.29.18325. [DOI] [PubMed] [Google Scholar]
- 5.Fairman JW, Noinaj N, Buchanan SK. Curr Opin Struct Biol. 2011;21:523–531. doi: 10.1016/j.sbi.2011.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Phillips R, Ursell T, Wiggins P, Sens P. Nature. 2009;459:379–385. doi: 10.1038/nature08147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schauer K, Rodionov DA, De Reuse H. Trends Biochem Sci. 2008;33:330–338. doi: 10.1016/j.tibs.2008.04.012. [DOI] [PubMed] [Google Scholar]
- 8.Postle K, Kadner RJ. Mol Microbiol. 2003;49:869–882. doi: 10.1046/j.1365-2958.2003.03629.x. [DOI] [PubMed] [Google Scholar]
- 9.Pierre JL, Fontecave M, Crichton RR. Biometals. 2002;15:341–346. doi: 10.1023/a:1020259021641. [DOI] [PubMed] [Google Scholar]
- 10.Wandersman C, Stojiljkovic I. Curr Opin Microbiol. 2000;3:215–220. doi: 10.1016/s1369-5274(00)00078-3. [DOI] [PubMed] [Google Scholar]
- 11.Wandersman C, Delepelaire P. Annu Rev Microbiol. 2004;58:611–647. doi: 10.1146/annurev.micro.58.030603.123811. [DOI] [PubMed] [Google Scholar]
- 12.Kotloff KL, Winickoff JP, Ivanoff B, Clemens JD, Swerdlow DL, Sansonetti PJ, Adak GK, Levine MM. Bull World Health Organ. 1999;77:651–666. [PMC free article] [PubMed] [Google Scholar]
- 13.Pazhani GP, Sarkar B, Ramamurthy T, Bhattacharya SK, Takeda Y, Niyogi SK. Antimicrob Agents Chemother. 2004;48:681–684. doi: 10.1128/AAC.48.2.681-684.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 15.Payne SM, Wyckoff EE, Murphy ER, Oglesby AG, Boulette ML, Davies NM. Biometals. 2006;19:173–180. doi: 10.1007/s10534-005-4577-x. [DOI] [PubMed] [Google Scholar]
- 16.Burkhard KA, Wilks A. J Biol Chem. 2007;282:15126–15136. doi: 10.1074/jbc.M611121200. [DOI] [PubMed] [Google Scholar]
- 17.Bracken CS, Baer MT, Abdur-Rashid A, Helms W, Stojiljkovic I. J Bacteriol. 1999;181:6063–6072. doi: 10.1128/jb.181.19.6063-6072.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cobessi D, Meksem A, Brillet K. Proteins. 2010;78:286–294. doi: 10.1002/prot.22539. [DOI] [PubMed] [Google Scholar]
- 19.Nepluev I, Afonina G, Fusco WG, Leduc I, Olsen B, Temple B, Elkins C. Infect Immun. 2009;77:3065–3074. doi: 10.1128/IAI.00034-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shen JS, Geoffroy V, Neshat S, Jia Z, Meldrum A, Meyer JM, Poole K. J Bacteriol. 2005;187:8511–8515. doi: 10.1128/JB.187.24.8511-8515.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yue WW, Grizot S, Buchanan SK. J Mol Biol. 2003;332:353–368. doi: 10.1016/s0022-2836(03)00855-6. [DOI] [PubMed] [Google Scholar]
- 22.Majumdar S, Hajduczki A, Mendez AS, Weiss GA. Bioorg Med Chem Lett. 2008;18:5937–5940. doi: 10.1016/j.bmcl.2008.07.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hajduczki A, Majumdar S, Fricke M, Brown IA, Weiss GA. ACS Chem Biol. 2011;6:301–307. doi: 10.1021/cb1001729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Majumdar S, Hajduczki A, Vithayathil R, Olsen TJ, Spitler RM, Mendez AS, Thompson TD, Weiss GA. J Am Chem Soc. 2011;133:9855–9862. doi: 10.1021/ja201792q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vithayathil R, Hooy RM, Cocco MJ, Weiss GA. J Mol Biol. 2011;414:499–510. doi: 10.1016/j.jmb.2011.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim KH, Aulakh S, Paetzel M. Protein Sci. 2012;21:751–768. doi: 10.1002/pro.2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.San-Miguel T, Perez-Bermudez P, Gavidia I. Springerplus. 2013;2:1–4. doi: 10.1186/2193-1801-2-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prive GG. Methods. 2007;41:388–397. doi: 10.1016/j.ymeth.2007.01.007. [DOI] [PubMed] [Google Scholar]
- 29.Hannavy K, Barr GC, Dorman CJ, Adamson J, Mazengera LR, Gallagher MP, Evans JS, Levine BA, Trayer IP, Higgins CF. J Mol Biol. 1990;216:897–910. doi: 10.1016/S0022-2836(99)80009-6. [DOI] [PubMed] [Google Scholar]
- 30.Kim J, Kapitan J, Lakhani A, Bour P, Keiderling TA. Theor Chem Acc. 2006;119:81–97. [Google Scholar]
- 31.Rossenu S, Leyman S, Dewitte D, Peelaers D, Jonckheere V, Van Troys M, Vandekerckhove J, Ampe C. J Biol Chem. 2003;278:16642–16650. doi: 10.1074/jbc.M208311200. [DOI] [PubMed] [Google Scholar]
- 32.Fusco WG, Choudhary NR, Council SE, Collins EJ, Leduc I. J Bacteriol. 2013;195:3115–3123. doi: 10.1128/JB.00199-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miles EW. Methods Enzymol. 1977;47:431–442. doi: 10.1016/0076-6879(77)47043-5. [DOI] [PubMed] [Google Scholar]
- 34.Chakraborti AS. Mol Cell Biochem. 2003;253:49–54. doi: 10.1023/a:1026097117057. [DOI] [PubMed] [Google Scholar]
- 35.Poulos TL. Chem Rev. 2014;114:3919–3962. doi: 10.1021/cr400415k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu X, Olczak T, Guo HC, Dixon DW, Genco CA. Infect Immun. 2006;74:1222–1232. doi: 10.1128/IAI.74.2.1222-1232.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith AD, Modi AR, Sun S, Dawson JH, Wilks A. Biochemistry. 2015;54:2601–2612. doi: 10.1021/acs.biochem.5b00017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Whitmore L, Wallace BA. Nucleic Acids Res. 2004;32:W668–W673. doi: 10.1093/nar/gkh371. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.