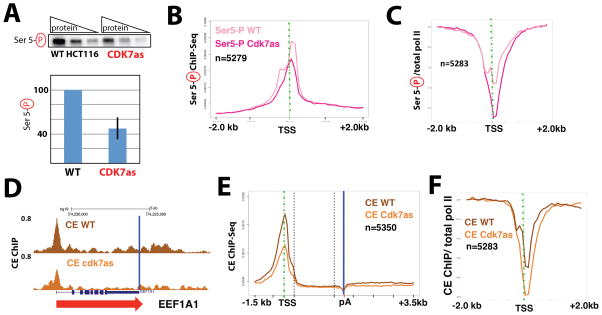
Figure 3.
Inhibition of CDK7 reduces CTD Ser5 phosphorylation and CE (RNGTT) recruitment. (A) Quantitative western data showing global phospho-Ser5 CTD levels in wild-type (WT) and CDK7as cells. Top: representative data with increasing total protein (3, 6, 12 micrograms); bottom: quantitation (± s.e.m) from n=3 technical replicates taken from nuclear extracts prepared from NM-PP1-treated WT or CDK7as cells, normalized to TBP. (B) Metaplots of mean anti-Pol II CTD Ser5-Phospho ChIP-Seq signals (normalized to yeast spike-in) in WT and CDK7as cells, each treated with NM-PP1 inhibitor (10 μM, 24h). Results are plotted for 100-base bins −2000 to +2000 bp from the TSS of highly expressed genes separated by over 2kb. (C) Metaplots of mean pol II CTD Ser5-Phospho ChIP signals normalized to total pol II in WT and CDK7as cells, each treated with NM-PP1, as in B. (D) UCSC genome browser screenshot of anti-capping enzyme (CE) ChIP-Seq signals at EEF1A1 gene in WT and CDK7as cells, each treated with NM-PP1, as in B. Blue vertical line corresponds to the poly(A) site. (E) Metaplots, as in B, for anti-CE ChIP signals in WT and CDK7as cells. 100-base bins are shown in flanking regions −1.5kb to +0.5kb relative to the TSS and −0.5kb to +3.5kb relative to poly(A) site. Gene body regions between +500 relative to the TSS and −500 relative to the poly(A) site were divided into 20 variable-length bins. (F) Metaplots of mean CE ChIP signals normalized to total pol II in WT and CDK7as cells, each treated with NM-PP1, as in B. See also Figure S3.