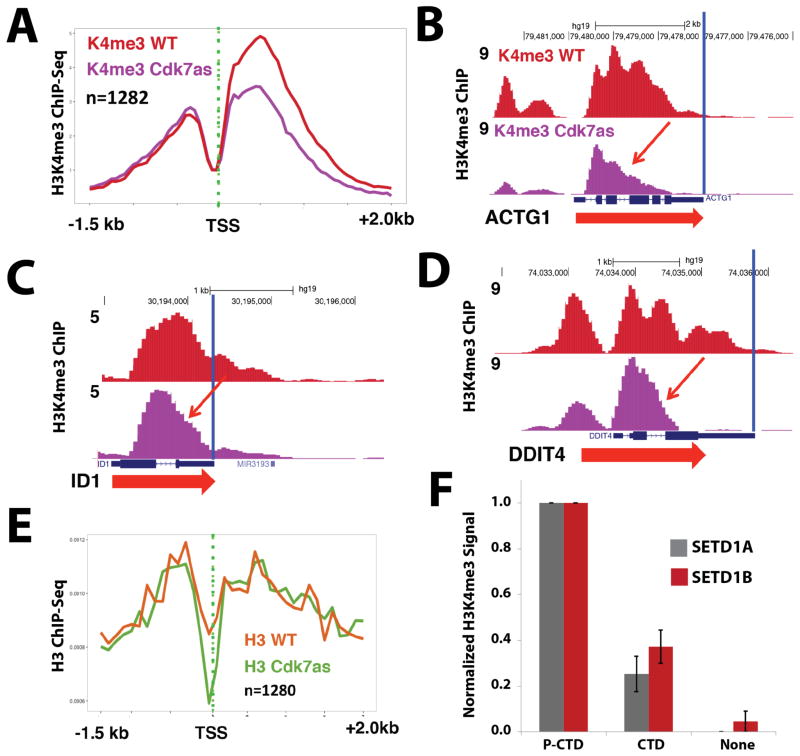
Figure 4.
CDK7 inhibition reduces H3K4me3 spreading; TFIIH-phosphorylated CTD stimulates SETD1A/B methyltransferase activity on nucleosomal templates. (A) Metaplots of mean anti-H3K4me3 ChIP-Seq signals in WT and CDK7as cells, each treated with NM-PP1 inhibitor (10 μM, 24h). Shown are data for a subset of genes with strongly reduced H3K4me3 signals upon CDK7 inhibition. (B–D) UCSC genome browser screen shots of anti-H3K4me3 ChIP-Seq signals in WT and CDK7as cells, each treated with NM-PP1 inhibitor, as in A. CDK7 inhibition limits H3K4me3 spreading into gene bodies (red arrows). (E) Metaplots of mean anti-H3 ChIP-Seq signals in WT and CDK7as cells for the subset of genes with strongly reduced H3K4me3 signals shown in panel A. (F) Quantitative western blots show elevated H3K4me3 levels when SETD1A or SETD1B enzyme reactions with nucleosomal substrates were supplemented with TFIIH-phosphorylated CTD vs. equivalent amounts of unmodified pol II CTD (none = no pol II CTD added). Data are from 3 biological replicates (± s.e.m.). See also Figure S3, S4, S5.