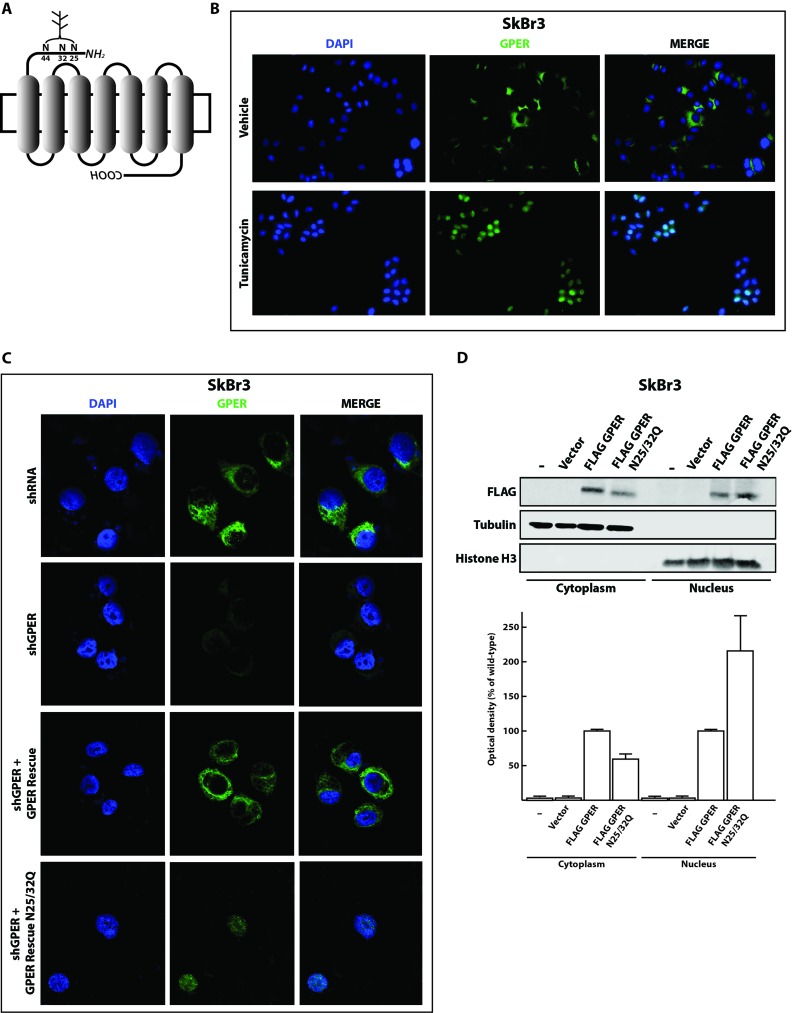
Figure 1. Inhibition of N-linked glycosylation induces the nuclear accumulation of GPER.
A. Schematic representation of the GPER protein structure. The asparagine residues N25, N32 and N44 are predicted N-linked glycosylation sites in the N-terminal portion of the receptor. B. Representative immunofluorescence micrographs of endogenous GPER in SkBr3 breast cancer cells treated or not for 24 hours with 5 μg/ml of tunicamycin and stained with an anti-GPER antibody (green staining). Nuclei were stained with DAPI (blue). Images shown are representative of 10 different random fields. C. GPER immunoflourescence micrographs of SkBr3 cells transfected for 24 hours with shRNA or shGPER constructs, or co-transfected, for additional 24 hours, with shGPER in combination with the shRNA-resistant GPER expression vector for either the wild-type GPER (GPER Rescue) or the N-glycosylation double mutant N25/32Q. Images shown are representative of 10 different random fields. D. Immunoblot of a subcellular fractionation experiment with SkBr3 breast cancer cells transfected as indicated. For these experiments, exogenous GPER is FLAG-tagged. Tubulin and histone H3 serve as markers and for standardization for the cytoplasmic and nuclear compartments, respectively. The bottom panel shows the densitometric analysis with each data point representing the mean ± SD of three independent experiments. After standardization to the respective compartmental markers, the values were expressed as % of the respective samples with FLAG-tagged wild-type GPER (each set to 100%).