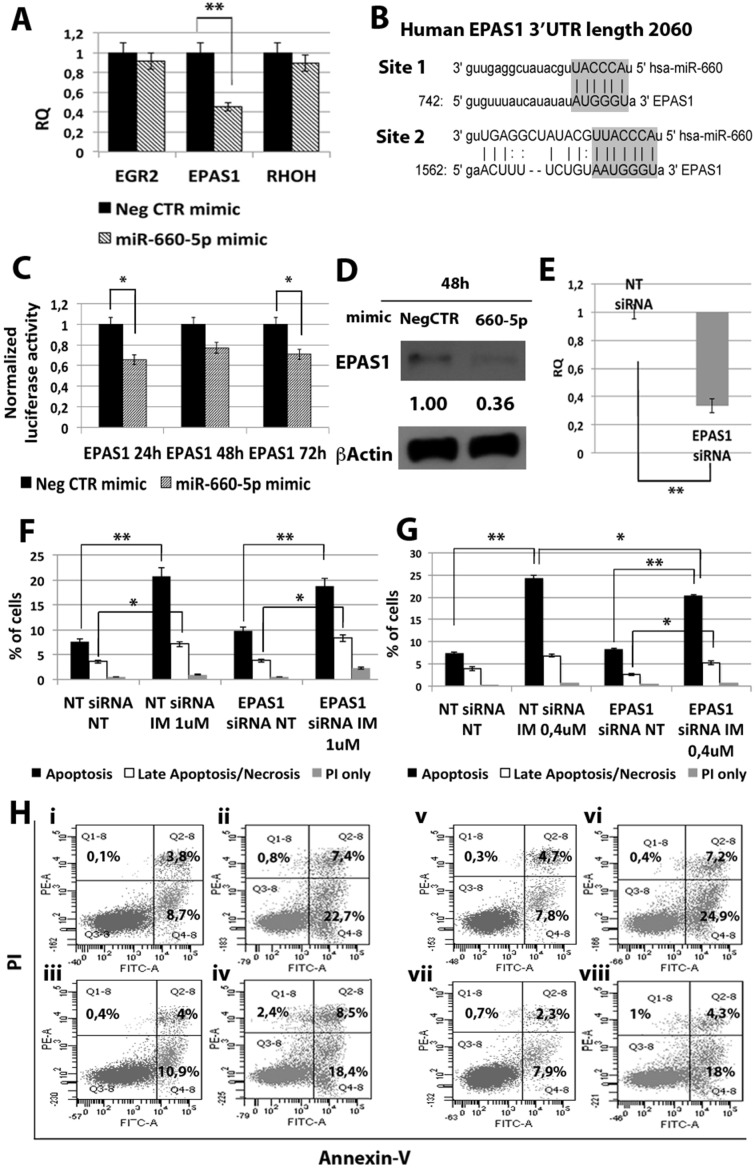
Figure 5. Effects of EPAS1 silencing on K562 cells' response to IM.
(A) qRT-PCR analysis of miR-660-5p predicted targets performed 24 h after miR-660-5p transfection. Results were normalized to the NegCTR mimic sample (n = 3). (B) Representation of the two miR-660-5p predicted binding sites in EPAS1 3′UTR sequence as reported by TargetScanHuman v7.0. The seed region of the miRNA is highlighted. (C) Normalized luciferase activity of K562 cells co-nucleofected with miR-660-5p miRNA mimic and EPAS1 3′UTR reporter vector. Each bar represents the luciferase activity upon miRNA overexpression normalized on the value of the same 3′UTR luciferase vector upon Neg-mimic transfection (set to 1). Values are reported as mean ± SEM (n = 3) *p < 0.05, **p < 0.01. (D) Western blot analysis of EPAS1 protein levels in whole cell lysates from K562 cells overexpressing miR-660-5p 48 hours after mimic nucleofection. EPAS1 protein level in miR-660-5p overexpressing cells was compared with control sample nucleofected with mimic Negative Control (Neg CTR). β-actin was included as loading control. (E) EPAS1 mRNA expression levels 24 hours after the last nucleofection as evaluated by qRT-PCR. Data are reported as RQ mean ± S.E.M of 3 independent experiments. Results of Annexin V/PI staining on K562 cells transfected with EPAS1 siRNA after 24 h (F) and 48 h (G) of IM treatment (mean ± SEM; n = 3). Representative dot plots for flow cytometry detection of Annexin V and PI staining at 24 h and 48 h after treatment are shown (H), i: NT siRNA NT, ii: NT siRNA IM 1 μM, iii: EPAS1 siRNA NT, vi: EPAS1 siRNA IM 1 μM, v: NT siRNA NT, vi: NT siRNA IM 0.4 μM, vii: EPAS1 siRNA NT, viii: EPAS1 siRNA IM 0.4 μM. *p < 0.05, **p < 0.01. Abbreviations: RQ, Relative Quantity; MIF indicates Mean Fluorescence Intensity; IM, Imatinib Mesylate; NT, Not Treated; 24 h, 24 hours; 48 h, 48 hours; PI, Propidium Iodide.