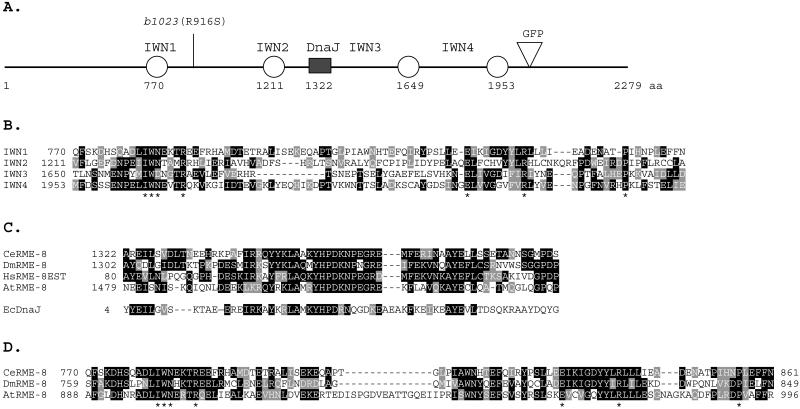
Figure 2.
RME-8 structure and sequence conservation. Conserved residues for sequence alignments (B–D) are shaded black and similar amino acids are shaded gray. (A) Domain structure of RME-8, showing a central DnaJ-domain (filled box) and four novel IWN repeats (circles). rme-8(b1023) has an amino acid substitution (R916S), as shown on top. In the RME-8::GFP fusion reporter, GFP (inverted triangle) is inserted after the fourth IWN repeat. (B) Alignment of the four IWN repeats in C. elegans RME-8. Invariant amino acids present in all IWN repeats are marked with asterisk at the bottom. These residues are also conserved in IWN repeats of other RME-8 homologues (see D for IWN1). (C) Sequence conservation among the J-domains of RME-8 homologues from C. elegans (CeRME-8), humans (HsRME-8EST), Drosophila (DmRME-8), and Arabidopsis (AtRME-8). The J-domain from E. coli DnaJ protein (EcDnaJ) is shown for comparison at the bottom. (D) Sequence conservation of the IWN1 region of RME-8 homologues.