Fig. 6.
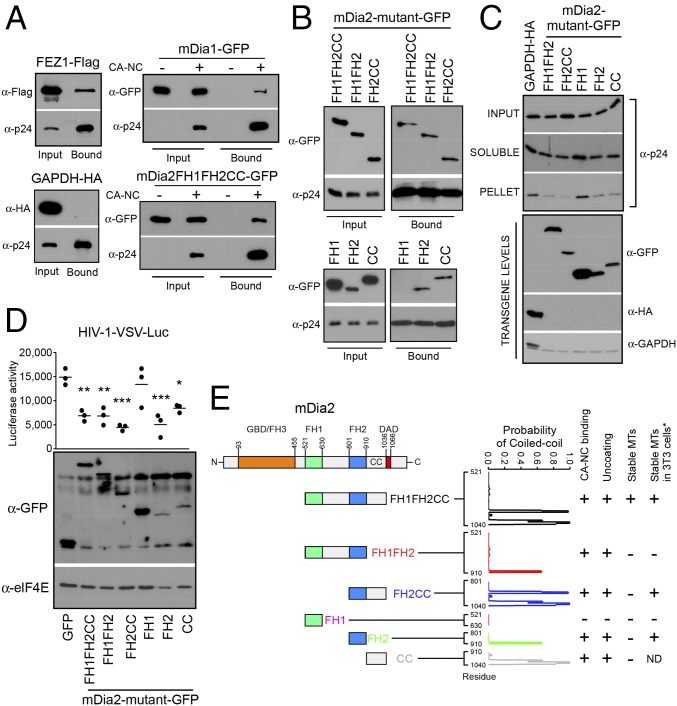
mDia interacts with in vitro assembled HIV-1 CA–NC complexes and regulates uncoating. (A and B) Binding of mDia to in vitro assembled HIV-1 CA–NC complexes. The 293T cells were transfected with plasmids expressing Flag-tagged FEZ1 (FEZ1-Flag) as positive control, HA-tagged GAPDH as negative control, and GFP-tagged mDia1 or mDia2 FH1FH2CC (A) or GFP-tagged truncation mutants of mDia2 containing FH1FH2CC, FH1FH2, FH2CC, FH1, FH2, and CC (B). Cells were lysed 36 h after transfection and lysates incubated at room temperature for 1 h with in vitro assembled HIV-1 CA–NC complexes. Samples were taken either before (input) or after sedimentation through a 70% sucrose cushion (bound) and analyzed by WB using indicated antibodies. (C) The 293A cells were transfected with control HA-tagged GAPDH or GFP-tagged forms of mDia2 consisting of FH1FH2, FH2CC, FH1, FH2, or CC domains. Cells were infected with HIV-1–VSV-Luc and the amounts of soluble and particulate capsid determined using the fate-of-capsid assay (Upper). Before infection, cells were lysed and analyzed by WB to determine transgene levels using anti-GFP, HA, and GAPDH antibodies (Lower). (D) CHME3 cells were transfected with plasmids expressing GFP control or GFP-tagged truncation mutants of mDia2: FH1FH2CC, FH1FH2, FH2CC, FH1, FH2, or CC. At 48 h posttransfection, cells were either infected with HIV-1–VSV–Luc followed by measurement of luciferase activity or lysed and subjected to WB analysis using anti-GFP and anti-elF4E (as loading control) antibodies. Data are shown as scatterplot with mean, n = 3. Statistical analysis was determined using one-way ANOVA with posttest, *P ≤ 0.05, **P ≤ 0.01, and ***P ≤ 0.001, respectively. (E) Schematic of the domains of mDia2 and domains studied (Left). Summary of whether each Dia2 construct binds CA–NC, affects uncoating, or induces stable MTs. * indicates results from previously published data (34) for comparison with our findings. The probability that each mutant will form a coiled coil is shown and was calculated using the COILS program, with the residue marked on the x axis.