Significance
Charging of tRNAs with the wrong amino acid can result in mistranslation of protein-encoding genes. Proofreading by tRNA editing domains clears these mischarged tRNAs, ensuring fidelity; however, structures of complexes with mischarged substrates are not currently available, and many mechanistic questions remain unanswered. ProXp-ala, present in all domains of life, selectively removes Ala from tRNAPro, leaving the cognate Pro-tRNAPro intact. A chemically synthesized nonhydrolyzable substrate analog has allowed characterization of the enzyme-substrate complex using NMR spectroscopy. Based on NMR studies, mutagenesis, enzymatic assays, molecular dynamics, and X-ray crystallography, we conclude that ProXp-ala uses multiple strategies, including conformational selection by a dynamic helix, size exclusion, and chemical discrimination, to ensure editing solely of Ala-tRNAPro.
Keywords: NMR, aminoacyl-tRNA synthetases, trans-editing, conformational selection, molecular dynamics
Abstract
Molecular sieves ensure proper pairing of tRNAs and amino acids during aminoacyl-tRNA biosynthesis, thereby avoiding detrimental effects of mistranslation on cell growth and viability. Mischarging errors are often corrected through the activity of specialized editing domains present in some aminoacyl-tRNA synthetases or via single-domain trans-editing proteins. ProXp-ala is a ubiquitous trans-editing enzyme that edits Ala-tRNAPro, the product of Ala mischarging by prolyl-tRNA synthetase, although the structural basis for discrimination between correctly charged Pro-tRNAPro and mischarged Ala-tRNAAla is unclear. Deacylation assays using substrate analogs reveal that size discrimination is only one component of selectivity. We used NMR spectroscopy and sequence conservation to guide extensive site-directed mutagenesis of Caulobacter crescentus ProXp-ala, along with binding and deacylation assays to map specificity determinants. Chemical shift perturbations induced by an uncharged tRNAPro acceptor stem mimic, microhelixPro, or a nonhydrolyzable mischarged Ala-microhelixPro substrate analog identified residues important for binding and deacylation. Backbone 15N NMR relaxation experiments revealed dynamics for a helix flanking the substrate binding site in free ProXp-ala, likely reflecting sampling of open and closed conformations. Dynamics persist on binding to the uncharged microhelix, but are attenuated when the stably mischarged analog is bound. Computational docking and molecular dynamics simulations provide structural context for these findings and predict a role for the substrate primary α-amine group in substrate recognition. Overall, our results illuminate strategies used by a trans-editing domain to ensure acceptance of only mischarged Ala-tRNAPro, including conformational selection by a dynamic helix, size-based exclusion, and optimal positioning of substrate chemical groups.
A fundamental function of the aminoacyl-tRNA synthetase (ARS) family of enzymes is to attach specific amino acids to cognate tRNA substrates for use in protein synthesis. Owing to the similar size of many amino acids, approximately one-half of the ARSs have evolved editing mechanisms that serve to clear misacylated tRNAs and maintain translational fidelity (1). Editing active sites, which are located in domains distinct from the amino acid activation domain, have been identified in both class I and class II synthetases. More recently, single-domain proteins that are homologous to class II synthetase editing domains also have been shown to clear misacylated tRNAs in trans (2–6).
Larger substrates are generally believed to be less problematic for ARSs than amino acids that are similar in size or smaller than the cognate substrate according to a proposed double-sieve mechanism (7, 8). Larger amino acids are rejected from the aminoacylation active site in the first sieve, whereas the second sieve accepts and clears the mischarged tRNA while rejecting the larger cognate substrate. In support of the sieve-based mechanism of discrimination, editing active sites present in both classes of ARSs have been shown to be tunable, with point mutations that alter the size of the amino acid binding pocket resulting in alternate substrate specificities (9–11). Paradoxically, Ser, which is larger than Ala, is misactivated by alanyl-tRNA synthetase (AlaRS) along with smaller Gly, and both Ser-tRNAAla and Gly-tRNAAla are edited by the AlaRS editing domain. In addition, a family of single-domain editing proteins known as AlaXps, which are homologs of the AlaRS editing domain, clear Ser-tRNAAla in trans (12). Rather than size-based exclusion, the AlaRS and AlaXp editing domains have been proposed to use chemical discrimination to reject Ala and accept only Ser via an active site Zn2+ ion (4, 13).
Single-domain editing enzymes in the INS superfamily are homologous to the prolyl-tRNA synthetase (ProRS) editing domain (INS). Some INS superfamily members function as a third sieve that deacylates mischarged tRNAs not cleared by ARS editing domains (3, 6, 14–16); for example, ProRS misactivates both Ala and Cys. Ala, which is smaller than cognate Pro, is hydrolyzed in the INS domain present in some ProRSs, whereas Cys, which is similar in size to cognate Pro, is cleared by a single-domain protein known as YbaK (3, 14, 15, 17). An alternative triple-sieve editing mechanism has been identified in some species that lack a ProRS editing domain but encode two free-standing editing domains, YbaK and ProXp-ala. The latter performs the same function as the INS domain, hydrolyzing Ala-tRNAPro, but solely in trans (6, 18). The mechanism of YbaK deacylation of Cys-tRNAPro has been well characterized and involves cyclization of the substrate sulfhydryl moiety (17); however, the structural and chemical basis of Ala recognition and Pro discrimination by ProXp-ala remain unknown.
We previously showed that Caulobacter crescentus (Cc) ProXp-ala achieves tRNAPro substrate specificity, in part through sequence determinants in the acceptor stem of tRNAPro, including recognition of the unique first base pair (C1:G72) (6, 18). Thus, Ala-tRNAAla, which lacks this element, is a poor substrate for ProXp-Ala, and binding by elongation factor-Tu (EF-Tu) further ensures that the cognate substrate is not deacylated (6, 18). How ProXp-ala avoids deacylation of Pro-tRNAPro is an open question. Ala has few distinguishing features for positive recognition, but several features of Pro may be used for negative discrimination, including its larger size, its unique ring structure, and the presence of a secondary α-amine group. To investigate the structural basis of substrate recognition by Cc ProXp-ala, we synthesized a nonhydrolyzable 3′-amide-linked mischarged acceptor stem mimic, nh-Ala-microhelixPro. Using NMR, we mapped the interactions between ProXp-ala and both uncharged microhelixPro and nh-Ala-microhelixPro. Furthermore, 15N-relaxation experiments identified a helix that exhibits fast ps-ns dynamics, as well as slower μs-ms motions, which are attenuated only when bound to the mischarged aminoacyl-tRNA mimic. These results, combined with extensive mutagenesis, deacylation assays, X-ray crystallography, and computational studies, reveal that a remarkable combination of substrate sampling, size-based selection, and specific chemical groups ensures rejection of cognate Pro-tRNAPro from the ProXp-ala active site.
Results
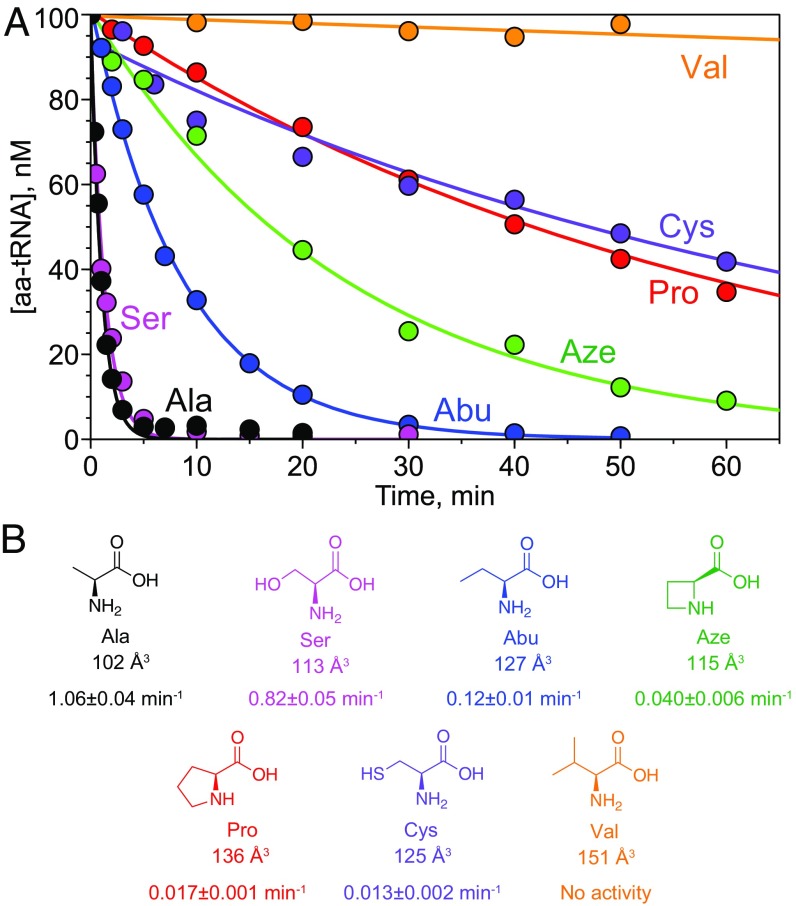
To test the role of size-based discrimination in the mechanism of substrate selection by Cc ProXp-ala, we assayed its ability to deacylate Escherichia coli (Ec) tRNAPro, a substrate for Cc ProXp-ala (6, 18), that was charged with amino acids of varying size, including Val, Pro, Cys, 2-aminobutyric acid (Abu), Ser, and azetidine-2-carboxylic acid (Aze), a smaller Pro analog with a four-membered ring (Fig. 1). With two exceptions, the measured kobs values are directly correlated with substrate aminoacyl moiety size, with smaller substrates generally demonstrating faster rates of deacylation under single-turnover conditions. Ala-tRNAPro is the best substrate, followed by Ser-, Abu-, Cys-, and Pro-tRNAPro. Val is the largest amino acid tested, and Val-tRNAPro deacylation was not detectable. Aze-tRNAPro exhibited a deacylation rate between that of Abu- and Pro- or Cys-tRNAPro, despite the fact that Aze is more similar in size to Ser, which was deacylated almost as efficiently as Ala (Fig. 1). Thus, both Cys- and Aze-tRNAPro were deacylated somewhat more slowly than expected based on size alone, suggesting that the chemical nature of the side chain plays a role as well.
Fig. 1.
Cc ProXp-ala deacylation of aminoacyl-tRNAPro. (A) Representative time courses for deacylation of 0.1 μM Ala-tRNAPro (black), Ser-tRNAPro (magenta), Aze-tRNAPro (green), Cys-tRNAPro (violet), Abu-tRNAPro (blue), Pro-tRNAPro (red), and Val-tRNAPro (orange) by 0.75 μM Cc ProXp-ala. (B) Chemical structures and molecular volumes of the aminoacyl moieties used in A. Observed rate constants of deacylation (kobs) are indicated below each structure. A representative time course is shown for each substrate, and kobs values are the average of three trials with the SD indicated.
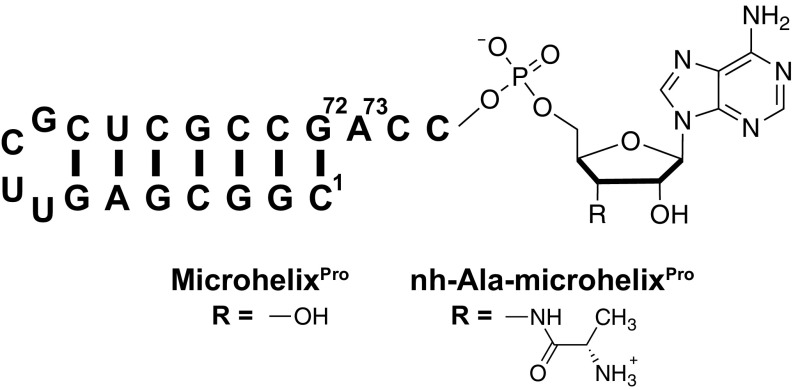
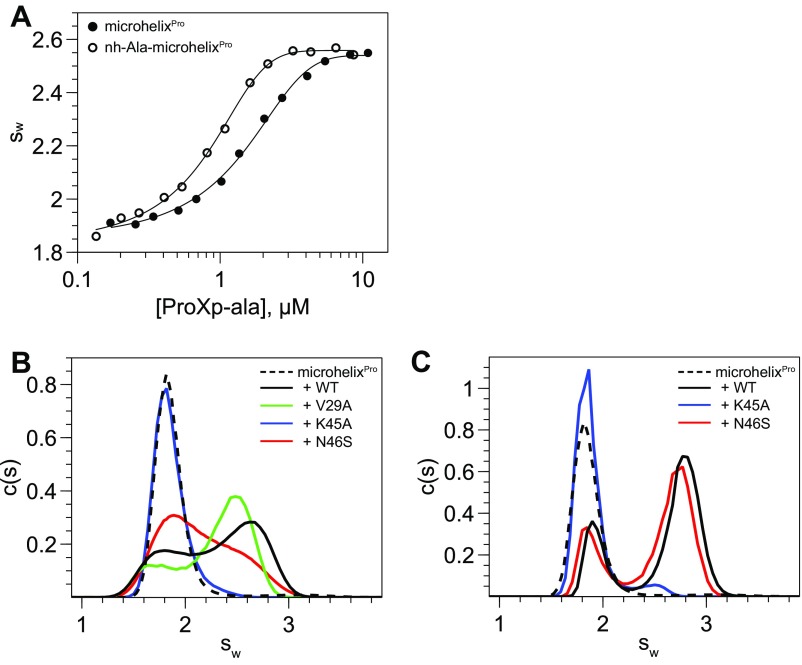
Cc ProXp-ala has previously been shown to deacylate an acceptor stem-derived Ala-microhelixPro substrate (Fig. 2), with only a twofold reduction in enzymatic activity (18). To investigate the mechanism of Ala-tRNA recognition by ProXp-Ala, we synthesized a nonhydrolyzable 3′-amide–linked nh-Ala-microhelixPro as a substrate analog, and used an uncharged microhelixPro to mimic the reaction product (Fig. 2). Apparent dissociation constants for ProXp-ala binding to microhelixPro and nh-Ala-microhelixPro, determined from sedimentation velocity analytical ultracentrifugation (AUC) experiments (19, 20), were 1.5 ± 0.3 μM and 0.52 ± 0.14 μM, respectively (Fig. S1A). Thus, the Ala moiety contributes modestly to enhancing the affinity of ProXp-ala for the misacylated substrate over the uncharged tRNA.
Fig. 2.
Ec tRNAPro-derived microhelix RNAs used in the study. MicrohelixPro is derived from the acceptor stem of Ec tRNAPro and contains a stable UUCG tetraloop. MicrohelixPro and nh-Ala-microhelixPro differ by the displayed “R” group; microhelixPro contains a free 3′ OH, whereas nh-Ala-microhelixPro contains an Ala moiety attached to the 3′ position via a nonhydrolyzable amide bond. The numbering of RNA bases refers to full-length Ec tRNAPro.
Fig. S1.
AUC data used for binding analyses in this study. (A) Isotherms of weight-average sedimentation coefficient as a function of ProXp-ala concentration. Analysis of the isotherm gives initial estimates of Kd values, which were refined by global data modeling using SEDPHAT. (B and C) Representative AUC sedimentation velocity data used to determine relative binding affinity of 1 μM microhelixPro to 1.2 μM (B) or 9.6 μM (C) WT or mutant ProXp-ala. Data are shown for microhelixPro alone (dashed line) and in the presence of WT (black), V29A (green), N46S (red), or K45A (blue) ProXp-ala. The c(s) distributions were integrated to find signal-weighted average sedimentation coefficients (sw). The sw values for each mutant were compared with that of 1.2 μM WT ProXp-ala (concentration at which ≈50% of microhelixPro is bound). The relative binding for each mutant was defined as follows: +++, equal or larger sw values for the 2.4 μM mutant, indicating a less than twofold reduction in affinity (as for V29A in B); ++, sw values smaller for the 2.4 μM mutant but larger for the 9.6 μM mutant, indicating a twofold to eightfold reduction in affinity (as for the N46S mutant in C); +, sw values smaller for the 9.6 μM mutant, indicating a greater than eightfold reduction in affinity; −, no binary complex observed up to the 9.6 μM mutant (as for K45A in C).
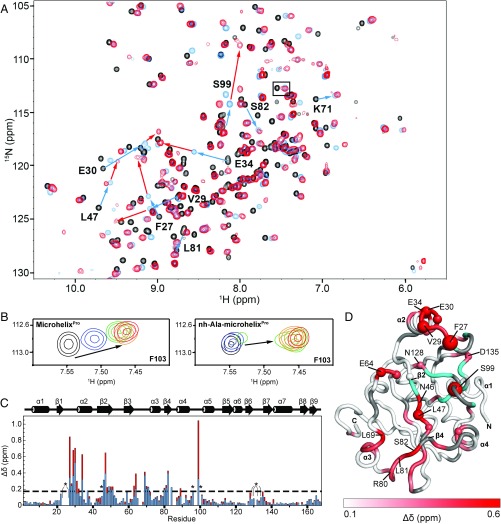
We used NMR spectroscopy to map the microhelixPro and nh-Ala-microhelixPro binding surfaces of Cc ProXp-ala. We first obtained backbone resonance assignments in the absence of substrates, and then recorded 2D 1H-15N HSQC spectra in the presence of varying concentrations of either microhelixPro or nh-Ala-microhelixPro (Fig. 3A). Resonance assignments were obtained for 156 of the 160 non-Pro residues (168 total) for free ProXp-ala, for 153 residues in the presence of microhelixPro, and for 140 residues in the presence of nh-Ala-microhelixPro; secondary chemical shifts (21) were generally consistent with the deposited crystal structure 1VJF (Fig. S2). Chemical shift perturbations (CSPs) were observed for a similar subset of residues of ProXp-ala with both microhelixPro and nh-Ala-microhelixPro, indicating a site-specific interaction with each RNA. Furthermore, the CSPs induced by Ala-microhelixPro are in the same direction as those for microhelixPro but are larger in magnitude (Fig. 3A).
Fig. 3.
15N-HSQC NMR spectra reveal similarities and differences in Cc ProXp-ala binding to microhelixPro or nh-Ala-microhelixPro. (A) 800 MHz 15N-HSQC spectrum of ProXp-ala alone (black) or in the presence of a 1:1 molar ratio of either microhelixPro (blue) or nh-Ala-microhelixPro (red). Blue arrows indicate signals with significant CSPs induced by microhelixPro, and red arrows indicate additional CSPs induced by nh-Ala-microhelixPro. Arrows are labeled with the corresponding residue of ProXp-ala. (B) Enlarged view of the regions of the 15N-HSQC spectrum boxed in A from the titrations of ProXp-ala with 0.2 (blue), 0.4 (green), 0.6 (orange), or 0.8 (red) equivalents of microhelixPro (Left) or nh-Ala-microhelixPro (Right). (C) Summary of per-residue CSPs [Δδ = (ΔδH2 + ΔδN2/25)1/2] on Cc ProXp-ala binding to microhelixPro (blue bars) or nh-Ala-microhelixPro (red bars). Secondary structure elements based on the Cc ProXp-ala crystal structure (PDB ID code 1VFJ) are displayed above the bar graph. The dashed line at 0.18 ppm represents the mean plus 1 SD of δΔ for microhelixPro. Residues whose amide resonances were broadened beyond detection are indicated by asterisks. (D) CSP values for nh-Ala-microhelixPro mapped onto the crystal structure as a linear gradient from white (≤0.1 ppm) to red (≥0.6 ppm). Prolines and other residues that could not be assigned in NMR spectra are in gray. Residues with peaks broadened beyond detection on the addition of nh-Ala-microhelixPro are colored cyan. Spheres are displayed for all residues with a CSP difference of at least 0.05 ppm, with relative sizes scaled to a range of 50–100% of the van der Waals radius.
Fig. S2.
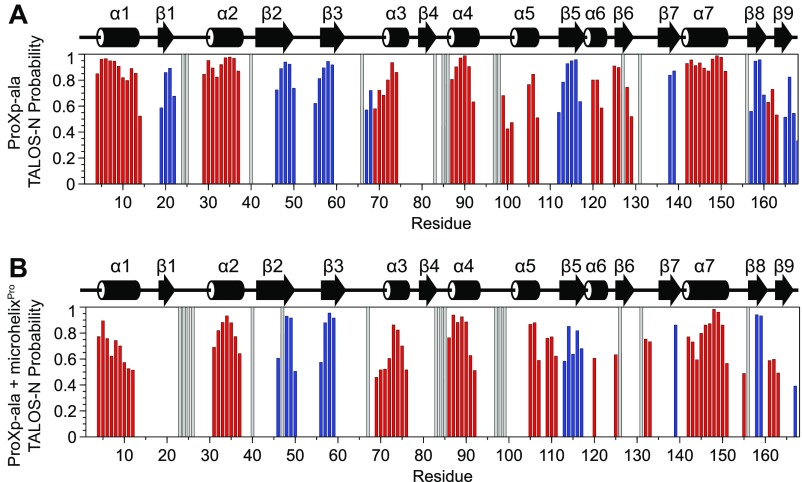
Secondary structure predictions from NMR chemical shift assignments. The TALOS-N webserver (https://spin.niddk.nih.gov/bax/nmrserver/talosn) was used to predict the secondary structures of free ProXp-ala (A) and ProXp-ala bound to microhelixPro (B) using assigned HN, N, Cα, and Cβ chemical shifts. The red bars indicate probability for the α-helix, and the blue bars indicate probability for the β-strand. Unassigned residues are depicted as gray bars. The secondary structure of the ProXp-ala crystal structure (PDB ID code 1VJF) is displayed above each plot for comparison.
NMR spectra of ProXp-ala recorded over a range of concentrations of uncharged microhelixPro revealed most of the perturbed backbone resonances to be in the fast exchange regime, characterized by an incremental shift in peak position with increasing RNA concentration (Fig. 3B). In contrast, titration with nh-Ala-microhelixPro revealed most backbone resonances of ProXp-ala to be in the intermediate-slow exchange regime, characterized by the broadening or disappearance of the resonance corresponding to its “free” position and reappearance of the same resonance in the “bound” position (Fig. 3B). This is consistent with higher affinity and/or slower off-rates for the mischarged substrate analog (Fig. S1A). Lineshape analysis (Fig. S3) of the NMR titration data for the uncharged microhelixPro yielded a Kd of 0.97 ± 0.21 μM, similar to that obtained from the AUC experiments, and a koff of 492 ± 28 s−1. Lower signal-to-noise, line broadening, and the slow/intermediate exchange behavior of the titration data with nh-Ala-microhelixPro resulted in greater uncertainty in the fitted Kd value (0.037 ± 0.04 μM), although better determined was a fivefold lower koff of 104 ± 8 s−1. Lineshape simulations indicated that intermediate-to-slow exchange is obtained with a Kd value consistent with that measured by AUC as long as koff is slower than ∼150 s−1.
Fig. S3.
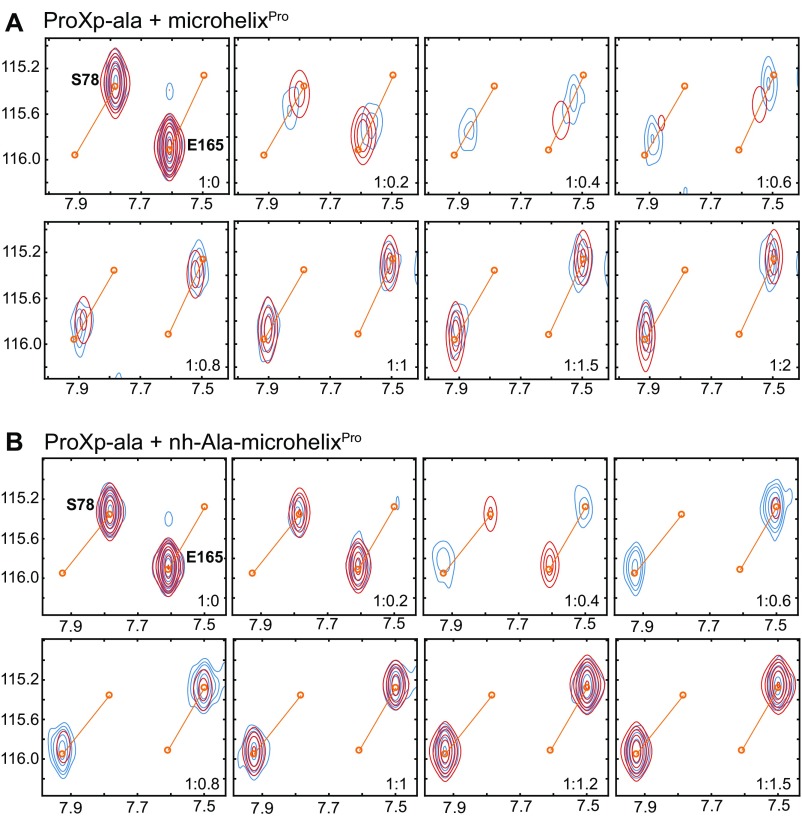
Representative results from 2D NMR lineshape analysis of Cc ProXp-ala bound to either microhelixPro (A) or nh-Ala-microhelixPro (B). Shown are experimental (blue) and fitted (red) contour plots and fitted free and bound peak positions (orange line) for each of the eight indicated titration points of residues S78 (Left) and E165 (Right). Lineshape analysis was conducted with TITAN (https://www.ucl.ac.uk/biosciences/departments/smb/christodoulou/titan) using a two-state binding model. Fitting of microhelixPro spectra (15 residues) yielded Kd = 0.97 ± 0.21 μM and koff = 492 ± 28 s−1, whereas fitting of nh-Ala-microhelixPro spectra (14 residues) yielded Kd = 0.037 ± 0.04 μM and koff = 104 ± 8 s−1.
Differences in CSPs between the substrate and product analogs pinpoint residues involved in recognition of the mischarged amino acid. In the presence of uncharged microhelixPro, significant CSPs (greater than the average CSP plus 1 SD) are observed for 20 residues of ProXp-ala (Fig. 3C, blue bars). Of the significant CSPs induced by nh-Ala-microhelixPro, 11 were similar to those induced by the uncharged RNA (e.g., K71, R80, L81, S82) (Fig. 3 A and C); however, some residues that showed weak to moderate CSPs in the presence of microhelixPro were significantly more perturbed by binding to nh-Ala-microhelixPro (F27, V29, E34, L47, E64, and S99) (Fig. 3 A and C). In addition to large perturbations throughout the enzyme, nine resonances assigned in the free protein were broadened beyond detection on the addition of nh-Ala-microhelixPro (H23, V26, K45, T96, V100, F129, H130, L132, and N134; Fig. 3C, asterisks). Overall, the greatest concentration of CSPs was seen in the region containing helix α2 and its preceding loop (residues 27–34). Significant perturbations are also observed for strand β2 (residues 46–51), the loop preceding helix α3 (residues 63–64 and 69–72), and strand β4 (residues 80–84). Residue S99 exhibits the largest overall perturbation and largest difference in CSP between the charged and uncharged substrates; assignments are not available for flanking residues (T96, P97, G98, and V100). Mapping the combined 1H and 15N CSPs for nh-Ala-microhelixPro (Fig. 3C) onto the crystal structure of Cc ProXp-ala reveals that all significant CSPs are localized to one face of ProXp-ala defined by α2, β2, and β4 (Fig. 3D). The resonances that were broadened beyond detection are located within a pocket on this same face of ProXp-ala (Fig. 3 C and D). CSP differences >0.05 ppm (Fig. 3D) include residues F27, V29, and E34 within helix α2 and its preceding loop, and residue S99. In addition, the residues adjacent to peaks broadened beyond detection on addition of nh-Ala-microhelixPro in the β6-β7 loop (N128 and D135) are more perturbed in the presence of the Ala moiety than with the uncharged microhelix. The electrostatic potential map of ProXp-ala (Fig. S4A) shows that these regions surround a large electropositive surface to complement the electronegative phosphate groups of the CCA-3′ end of the tRNA.
Fig. S4.
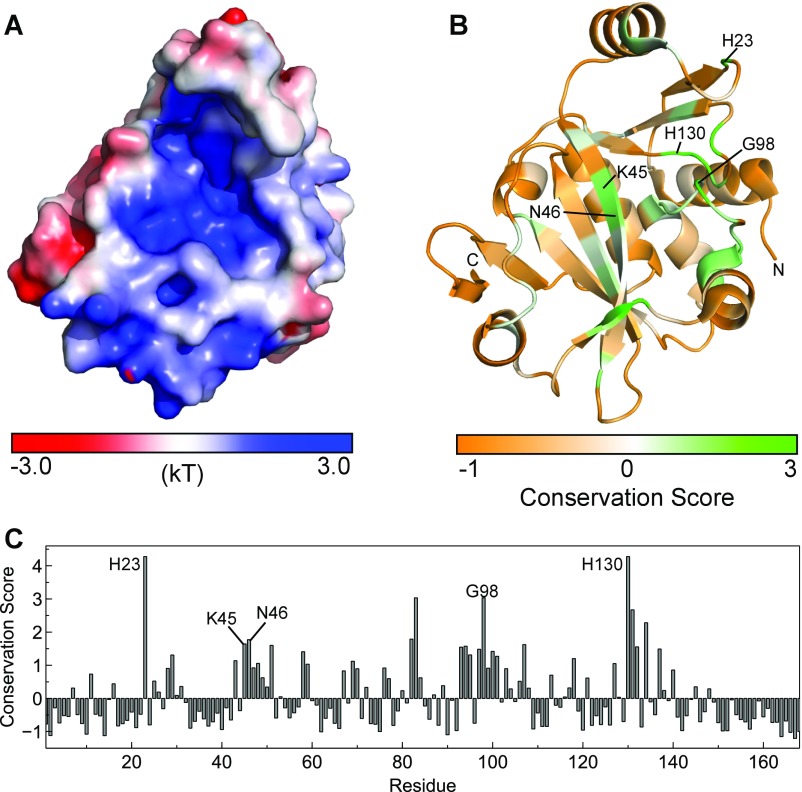
Electrostatic potentials and conservation of Cc ProXp-ala. (A) Electrostatic potentials of Cc ProXp-ala plotted on the molecular surface (PDB ID code 1VJF) as a gradient from red (−3.0 kT) to white (0 kT) to blue (3.0 kT). Potentials were calculated using the adaptive Poisson–Boltzmann solver (APBS) plugin within PyMOL. (B) Normalized conservation score within all Bacteria of Cc ProXp-ala plotted on the structure (PDB ID code 1VJF) as a gradient from orange (−1) to white (0) to green (3). Scores were determined from the AL2CO webserver (prodata.swmed.edu/al2co/al2co.php). (C) Graph of normalized conservation scores used to generate the map in B. The highly conserved residues mentioned in this study are labeled in B and C.
Guided by the CSPs (Fig. 3) and sequence conservation (Fig. S4 B and C), we generated site-directed mutants (to Ala) to examine effects on substrate binding and catalysis (Table 1). We performed deacylation assays with full-length Ec Ala-tRNAPro as the substrate and AUC-binding assays using microhelixPro (Fig. S1 B and C). Mutations in helix α2 and the preceding β1-α2 loop resulted in only minor decreases in deacylation (≤3.5-fold) and binding (≤8-fold), with the exception of highly conserved H23, which showed a 12-fold decrease in activity. Mutation of H23 and α2 residue E34 resulted in significantly (>8-fold) reduced binding. No deacylation activity or binding was detected for the K45A variant, consistent with the critical role of this β2 residue in positioning the CCA-3′ end into the active site, as previously reported for the INS domain of ProRS and YbaK (9, 17, 22–24). Mutation of the β2 residues H43 to Ala and N46 to Ser (N46A could not be purified) also showed significantly reduced deacylation activity (by 8- and 14-fold, respectively) and 2- to 8-fold reduced binding. S99 displayed the largest CSP difference in the presence of nh-Ala-microhelixPro, but resulted in only minor (∼3-fold) defects in deacylation and binding on mutation. In contrast, mutation of G98 had a large effect on enzyme activity (95-fold), but only a modest effect on binding (Table 1). Although unassigned in our NMR experiments, G98 is part of a highly conserved GXXXP motif in INS-like domains and has been proposed to be involved in stabilization of the transition state during catalysis (25). The N128A, H130A, and N134A mutants demonstrated moderate decreases in deacylation (6.3- to 11-fold), whereas V127A ProXp-ala was only 2-fold less active than wild type (WT). V127A, N128A, and H130A resulted in weaker binding, but N134A had no observable binding defect, despite an 11-fold decreased rate of deacylation. Thus, mutagenesis experiments suggest that the side chains of conserved residues within β2, the α4-α5 loop, and the β6-β7 loop are generally more critical for both binding and catalysis than the residues within helix α2 and the β1-α2 loop.
Table 1.
WT and mutant Cc ProXp-ala deacylation rate constants and relative Ec microhelixPro binding affinity
| Mutation | Location | Deacylation kobs, min−1* | Fold decrease | Relative binding† |
| WT | 1.05 ± 0.07 | 1 | +++ | |
| H23A | β1-α2 | 0.086 ± 0.019 | 12 | + |
| P24A | β1-α2 | 1.07 ± 0.07 | 0 | +++ |
| P25A | β1-α2 | 0.75 ± 0.11 | 1.4 | +++ |
| F27A | β1-α2 | 0.35 ± 0.04 | 3.0 | ++ |
| R28A | β1-α2 | 0.30 ± 0.04 | 3.5 | ++ |
| V29A | α2 | 0.53 ± 0.11 | 2.0 | +++ |
| E30A | α2 | 2.2 ± 0.2 | 0 | +++ |
| E31A | α2 | 0.29 ± 0.04 | 3.6 | +++ |
| E34A | α2 | 0.54 ± 0.05 | 2.0 | + |
| H43A | β2 | 0.13 ± 0.02 | 8.2 | ++ |
| K45A | β2 | ND | >100 | − |
| N46S | β2 | 0.076 ± 0.007 | 14 | ++ |
| G98A | α4-α5 | 0.011 ± 0.002 | 95 | ++ |
| S99A | α4-α5 | 0.39 ± 0.10 | 2.7 | ++ |
| V127A | β6 | 0.50 ± 0.06 | 2.1 | ++ |
| N128A | β6 | 0.17 ± 0.01 | 6.3 | + |
| H130A | β6-β7 | 0.11 ± 0.03 | 9.7 | + |
| N134A | β6-β7 | 0.098 ± 0.02 | 11 | +++ |
ND = not detected.
Deacylation rate constants (kobs) were determined using 0.75 μM enzyme and 0.1 μM Ala-tRNAPro at 20 °C. All results are an average of three trials with the SD indicated.
AUC sedimentation velocity experiments were used to determine the relative binding affinity of microhelixPro to mutant ProXp-ala, as described in Fig. S1 B and C. Binding relative to WT ProXp-ala is indicated as follows: +++, less than twofold reduction; ++, twofold to eightfold reduction; +, greater than eightfold reduction; −, no observed binding.
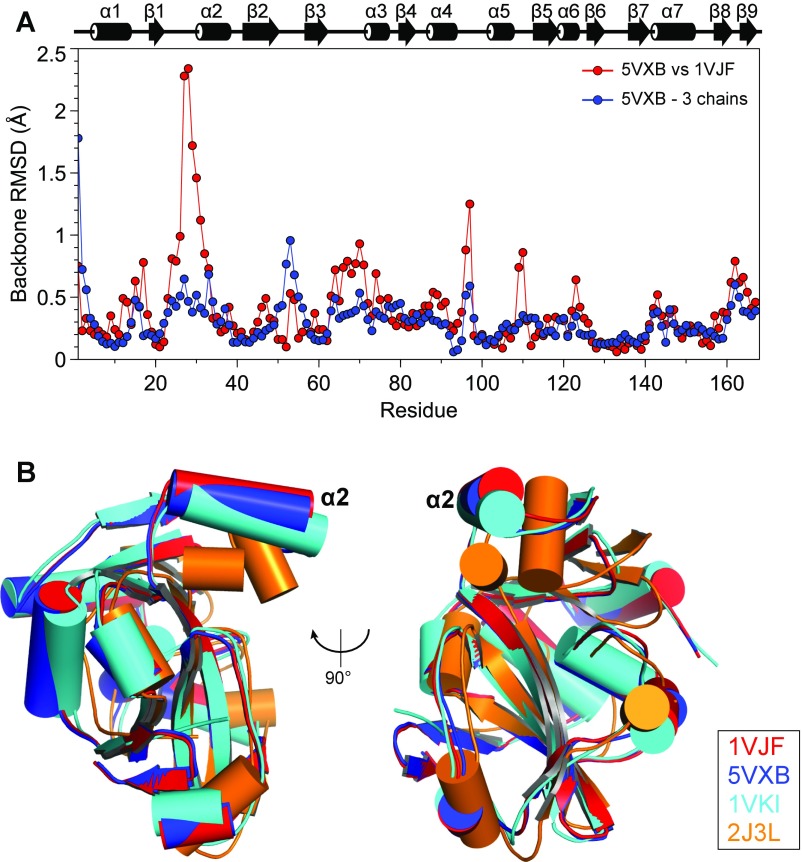
Comparison of the crystal structures of ProXp-ala domains to the INS domain from Enterococcus faecalis ProRS [Protein Data Bank (PDB) ID code 2J3L; residues 233–402] (9, 22, 25) suggests that a local conformational change on substrate binding may be responsible for the CSPs observed in helix α2 (Fig. S5). The ProXp-ala domains from Cc and Agrobacterium tumefaciens (At; PDB ID code 1VKI) share 43% sequence identity and have nearly identical structures, with a backbone atom rmsd of 1.11 Å. INS shares 12% and 11% sequence identity with Cc and At ProXp-ala, respectively (22). In addition, we independently determined the crystal structure of Cc ProXp-ala at 1.69-Å resolution in a new space group (PDB ID code 5VXB; Table S1) and found its structure to be nearly identical to that of 1VJF (rmsd <0.5 Å), except for the β1-α2 loop and the beginning of helix α2 (rmsd 1–2.5 Å) (Fig. S5A). In a superposition of each of these structures, the most striking difference is in the orientation of helix α2 between the INS domain and the three trans-editing domains (Fig. S5B). Based on this observation, we hypothesize that helix α2 may dynamically sample “open” and “closed” conformations.
Fig. S5.
Comparison of the crystal structures of ProXp-ala and homologs. (A) Backbone RMSD values per residue between the two crystal structures of Cc ProXp-ala (PDB IDs 1VJF and 5VXB) and average backbone rmsd values per residue among the three monomers in the asymmetric unit of 5VXB. A secondary structure representation is displayed above the plot. (B) Superposition of Cα atoms of Cc ProXp-ala (red, PDB ID code 1VJF; blue, PDB ID code 5VXB), At ProXp-ala (cyan; PDB ID code 1VKI), and Ef ProRS INS (orange, PDB ID code 2J3L; residues 233–402). The alpha helix corresponding to Cc helix α2 is labeled.
Table S1.
X-ray crystallography data collection and refinement statistics for Cc ProXp-ala
| Variable | Value* |
| Space group | P1211 |
| Cell parameters | |
| a, b, c, Å | 62.04, 67.73, 62.93 |
| α, β, γ, ◦ | 90.00, 118.62, 90.00 |
| Resolution (Å) | 19.80–1.69 (1.78–1.69) |
| Average redundancy | 6.7 (6.4) |
| Average I/σ(I) | 16.3 (3.3) |
| Rp.i.m. | 0.026 (0.228) |
| Completeness, % | 99.6 (99.0) |
| Refinement statistics | |
| No. of reflections | 51,071 (7,401) |
| R factor | 0.1664 |
| Free R factor | 0.1896 |
| Free R percentage | 4.01 |
| Average B factor, Å2 | 25 |
| rmsd from ideality | |
| Bond lengths, Å | 0.37 |
| Bond angles, ◦ | 0.57 |
| Ramachandran favored, % | 99 |
| Ramachandran allowed, % | 1 |
| Ramachandran outliers, % | 0 |
Values in parentheses are for the highest-resolution shell.
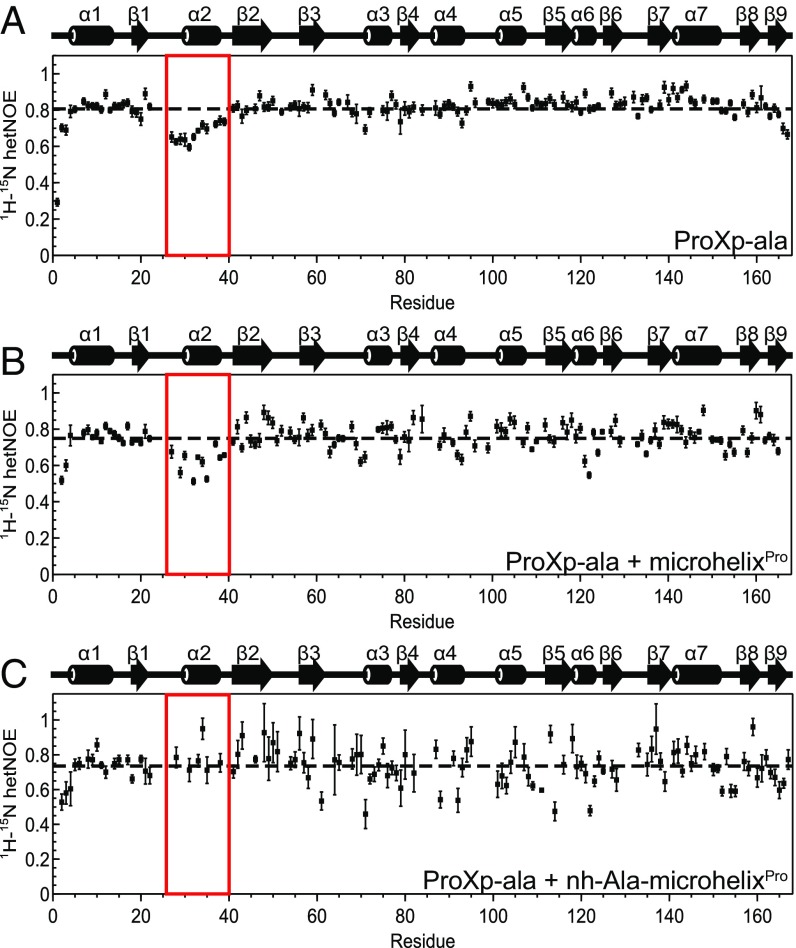
To test this conformational sampling hypothesis, we first used 15N NMR relaxation measurements to characterize the ps-ns dynamics of Cc ProXp-ala (26–28). Steady-state 1H-15N heteronuclear NOE values could be measured for 136 of the 168 residues of ProXp-ala (Fig. 4A). 1H-15N NOE values were relatively uniform, with an average of 0.81, except for the N and C termini and for residues 27–39, corresponding to helix α2 and the β1-α2 loop (Fig. 4A). The values observed in these residues indicate significant flexibility on the ps-ns timescale. These ps-ns dynamics persist for the same set of residues when ProXp-ala is bound to uncharged microhelixPro, as evidenced by steady-state 1H-15N heteronuclear NOE values (Fig. 4B). Although the lower signal-to-noise ratios in the protein-RNA complexes limit quantitative analysis, on binding to nh-Ala-microhelixPro, 1H-15N heteronuclear NOE values for the β1-α2 loop and helix α2 become uniform with the rest of the protein, indicating attenuation of ps-ns internal motions (Fig. 4C). Because the amide 1H-15N heteronuclear NOE is dominated by motions on the time scales of the 1H and 15N Larmor frequencies (29), these observations indicate fast ps-ns motions in the β1-α2 loop and helix α2.
Fig. 4.
Amide 15N-relaxation values indicate ps-ns dynamics in α2 of Cc ProXp-ala, which are attenuated on substrate recognition. (A) 1H-15N heteronuclear NOE data for 0.5 mM free ProXp-ala. (B) 0.5 mM ProXp-ala bound to a molar equivalent of microhelixPro. (C) 0.2 mM ProXp-ala bound to a molar equivalent nh-Ala-microhelixPro. The red box in each indicates stretch of residues near helix α2 with values lower than the average (dashed line) in free ProXp-ala. Secondary structure elements of ProXp-ala are displayed above the plots.
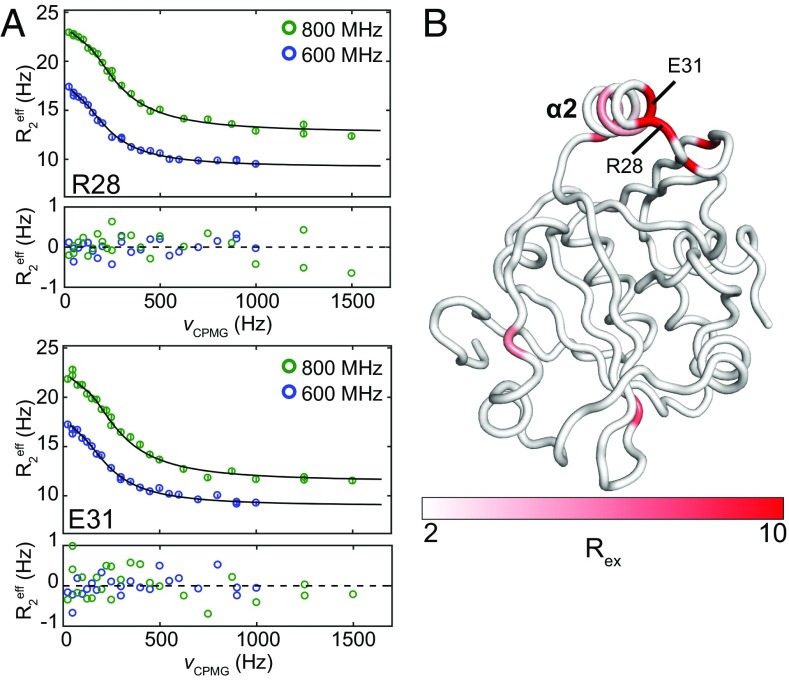
Because concerted conformational changes in helix α2 imply motions on timescales slower than ps-ns, we performed 15N-single quantum Carr–Purcell–Meiboom–Gill (CPMG) relaxation dispersion NMR experiments (30) to characterize chemical exchange on the μs-ms time scale (Fig. 5A). Of the 119 residues for which relaxation curves could be determined and assigned, 14 exhibit significant exchange on this time scale (Rex >2 s−1). Nine of the 14 localize to helix α2 and the β1-α2 loop (D22, V26, R28, E31, G32, L33, I35, A38, and M39), and the other five are distributed throughout the enzyme (L56, D68, R80, S99, and V140) (Fig. 5B). The nine residues in helix α2 and the β1-α2 loop fit well to a two-state exchange model, yielding a global exchange rate (kex) of 954 ± 57 s−1 and a major state population (PA) of 98.3% ± 0.1%. These observations are consistent with a model in which conformational sampling by residues around helix α2 enables substrate binding and selectivity.
Fig. 5.
Amide 15N CPMG relaxation dispersion experiments reveal μs-ms exchange dynamics involving helix α2 of Cc ProXp-ala. (A) Relaxation dispersion curves for ProXp-ala residues R28 and E31 at 600 and 800 MHz globally fit to a two-state exchange model with a kex of 950 s−1. Residuals of the fit are plotted below each graph. (B) Exchange contributions to transverse relaxation, Rex, mapped to the structure as a linear gradient from white (≤2 s−1) to red (≥10 s−1).
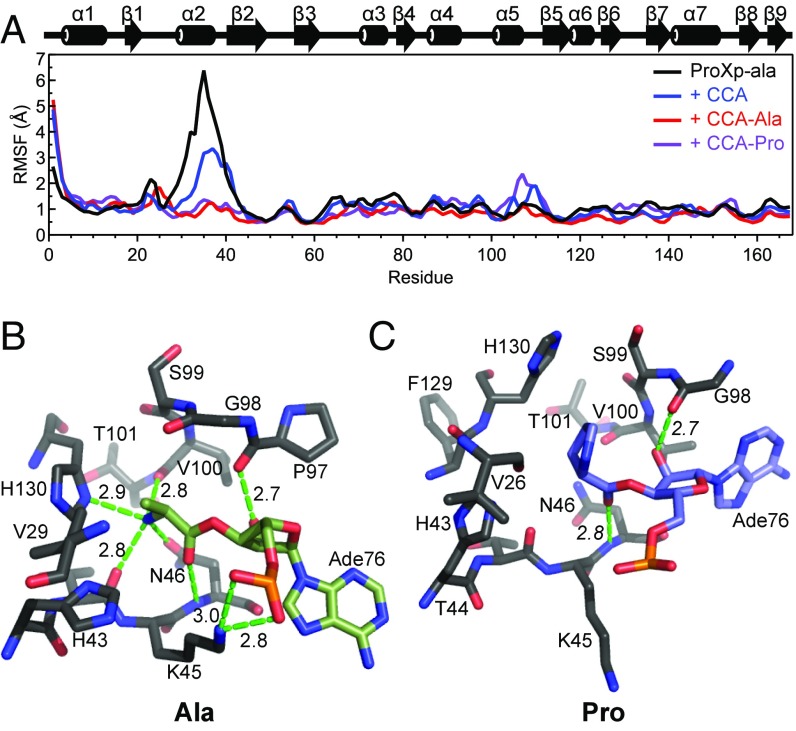
To further explore the dynamic sampling hypothesis, we used molecular dynamics (MD) simulations to investigate the conformational flexibility of helix α2 of Cc ProXp-ala in its free and ligand-bound states. To simulate the closed conformation, we built a homology model using the INS domain from Ef ProRS (PDB ID code 2J3L; residues 233–402; Fig. S5B) as a template (9, 22, 25). With this model, we performed docking simulations using the terminal portion of the Ala-tRNAPro ligand (CCA-Ala), such that the ligand was flexible and the Cc ProXp-ala active site side chains were allowed to sample alternative rotamer conformations. Out of 2,000 successful poses, the largest ligand cluster (pairwise rmsd of Ala heavy atoms <1 Å) featured Ala moiety binding in the active site pocket, with the positioning of the remaining RNA nucleotides highly variable. From the cluster, a representative pose was used as the starting point for a 15-ns explicit solvent MD simulation to allow the ligand to sample a small range of conformational space. The resulting structure was used as the starting model for subsequent MD simulations in which the aminoacyl moiety was modified to Pro to mimic properly charged tRNA (ProXp-ala + CCA-Pro), or the aminoacyl moiety was deleted to mimic the product (ProXp-ala + CCA). All analyses were performed on 40 ns of production simulation after equilibration. To simulate the dynamics of the unbound protein, we extended a trajectory from a ProXp-ala + CCA MD simulation in which the CCA ligand dissociated after ∼42 ns. In these simulations, the coordinate root mean square fluctuations (RMSFs) showed helix α2 to be significantly more flexible in the free protein but with reduced dynamics when bound to the CCA-aminoacyl ligand. Interestingly, the presence of either aminoacyl group (Ala or Pro) reduced the dynamics similarly (Fig. 6A).
Fig. 6.
RMSFs and active site snapshots from MD simulations. (A) RMSF in backbone nitrogen atom coordinates for each residue of ProXp-ala either alone (black) or in the presence of a CCA trinucleotide (blue), CCA-Ala (red), or CCA-Pro (purple) over a 40-ns MD simulation trajectory. Secondary structure elements of ProXp-ala are displayed above the plot. (B and C) Substrate conformations in the Cc ProXp-ala active site after flexible docking and MD simulations for “mischarged” CCA-Ala (B, green) and cognate CCA-Pro (C, blue). A76-aa is shown in each case, and only ProXp-ala residues within at least 5 Å of the aminoacyl moiety are shown. Potential hydrogen bond distances are indicated in Å.
Investigation of the ligand-bound active site after 30 ns of MD simulation revealed a significant difference between the CCA-Ala and CCA-Pro substrates (Fig. 6 B and C). The CCA-Ala ligand is stabilized by the potential for eight hydrogen bonds, five with the Ala moiety and an additional three with the backbone of A76 (Fig. 6B). In the presence of CCA-Pro, the ligand is shifted slightly out of the active site, correlating with a shift in the position of H130 toward the GXXXP loop (Fig. 6C). The GXXXP loop is also significantly perturbed when CCA-Pro is bound. The altered conformation of H130, the GXXXP loop, and the ligand cause a rotation in the A76 backbone, resulting in only transient interactions between the phosphate of A76 and K45, as well as the potential for only two intermolecular hydrogen bonds to stabilize the interaction.
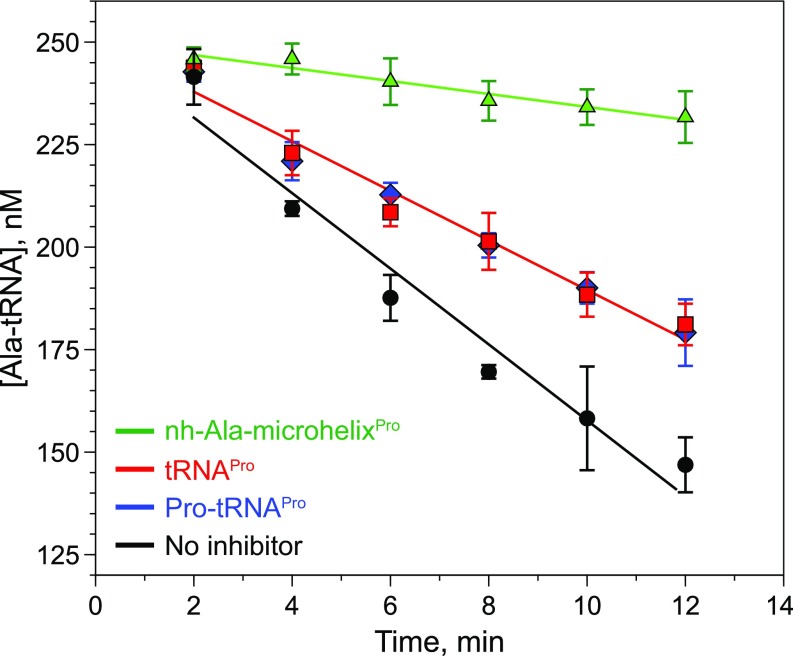
Our MD results are consistent with stabilization of the closed conformation of helix α2 in the presence of CCA-Ala, but we found a similar result for CCA-Pro (Fig. 6A). However, the significant distortion of the active site suggests that CCA-Pro is unlikely to bind efficiently or in a catalytically productive manner (Fig. 6C). Therefore, we tested whether cognate Pro-tRNAPro could inhibit Ala-tRNAPro deacylation and thus compete for binding to ProXp-ala. Our results show that Pro-tRNAPro is a very poor inhibitor of deacylation, behaving just like uncharged tRNA, demonstrating that Pro does not make any significant contribution to overall Pro-tRNAPro binding (Fig. S6). In contrast, nh-Ala-microhelixPro is a potent competitor, substantially reducing the rate of deacylation of Ala-tRNAPro when present at a fourfold excess over the cognate substrate, further validating its use in this study (Fig. S6).
Fig. S6.
Inhibition of Cc ProXp-ala deacylation of Ala-tRNAPro. Shown is the fitting of time courses for deacylation of 250 nM Ala-tRNAPro by 20 nM Cc ProXp-ala in the absence of an inhibitor (black circles) or in the presence of 1 µM Pro-tRNAPro (blue diamonds), 1 µM tRNAPro (red squares), or 1 µM nh-Ala-microhelixPro (green triangles). All results are the average of three trials with SDs indicated.
Discussion
Trans-editing domains are believed to be ancient proteins that preceded the addition of homologous domains into tRNA synthetases (31, 32). Here we show that the bacterial trans-editing enzyme ProXp-ala discriminates Ala-tRNAPro from Pro-tRNAPro through various strategies, including conformational selection in the substrate binding site, size-based exclusion of nonsubstrates, and chemical discrimination. Conformational differences in the position of active site-flanking helix α2 between crystal structures of ProXp-ala and INS suggest that it might sample both open and closed conformations (Fig. S5). Consistent with this hypothesis, our NMR relaxation experiments showed that in the free protein, helix α2 is dynamic across multiple timescales (ps-ns and μs-ms) (Figs. 4A and 5). NMR resonances for residues proximal to this region showed greater CSPs in the presence of the Ala moiety relative to the uncharged microhelixPro, suggesting stabilization of a closed conformation on binding of the aminoacylated substrate. Likewise, 1H-15N heteronuclear NOE experiments showed attenuation of fast-timescale dynamics only when the tRNA mimic was charged with the Ala moiety. These observations support a model in which ProXp-ala uses conformational selection as a mechanism of ligand sampling and discrimination.
ProXp-ala catalyzes the same reaction as the homologous INS editing domain present in many bacterial ProRSs. In the Ef ProRS INS domain, the backbone carbonyl of G261 (V26 in the β1-α2 loop of Cc ProXp-ala) and the 2′OH of the substrate A76 are proposed to hydrogen bond with a water, activating it for nucleophilic attack on the scissile ester carbon. Proton transfer from the water to the O3′ of A76 results in cleavage of the 3′O-linked ester bond (25). In the case of ProXp-ala, a mechanism involving backbone and tRNA functional groups is supported by the fact that, with the exception of K45, which has a proposed role in CCA-3′ end substrate positioning (9, 17, 22–24), the activity of the trans-editing domain is not dramatically affected on mutation of any other active site residues (Table 1). In addition, V26 is broadened beyond detection in the presence of nh-Ala-microhelixPro (Fig. 3C), supporting its proximity to the substrate.
The deacylation assays show a strong inverse correlation between hydrolysis rate and volume of the amino acid. This correlation is imperfect, however, because Aze and Ser have similar volumes but Ser is hydrolyzed 20 times faster, and because Cys and Abu are of similar size but Cys hydrolysis is only 1/10th as efficient as that of Abu. The cyclic rings of Pro and Aze both feature secondary amines, whereas the thiol group on Cys distinguishes it structurally and electronically from Ser or Abu. These observations reveal likely contributions from conformational and electronic features of the potential substrates. Interestingly, although extant ProRS is not known to misactivate Ser, we found that Ser-tRNAPro is hydrolyzed as efficiently as Ala-tRNAPro, suggesting that this activity of Cc ProXp-ala may be an evolutionary remnant.
Computational docking experiments provided useful insight into the structural basis for discrimination. The CCA-Ala-docked active site model contains numerous potential hydrogen bonds between active site residues and the primary α-amine group of Ala (Fig. 6B). Side chains of N46 and H130 are each within hydrogen-bonding distance, consistent with significantly decreased activity upon mutation (Table 1). Backbone carbonyls of T44 and V100 also are within hydrogen-bonding distance, consistent with CSPs induced by nh-Ala-microhelixPro. Residue K45 is universally conserved among INS superfamily members with one exception, and has been shown to play a role in substrate positioning (9, 17, 22–24). In our active site model, the side chain of K45 is within hydrogen-bonding distance of the A76 phosphate group, in agreement with those findings. In contrast to CCA-Ala, low-energy models of CCA-Pro docked into the active site feature a Pro ligand rotated around the ester bond (A76O2′-C) and around the C-Cα bond, such that the secondary α-amine group is not positioned for favorable hydrogen bonds (Fig. 6C). The lack of favorable contacts is consistent with the poor Pro-tRNAPro inhibition of Ala-tRNAPro deacylation observed experimentally (Fig. S6).
These studies were enabled by the availability of a stable aminoacyl-tRNA acceptor stem substrate mimic. A significant obstacle to the structural determination of enzyme-aminoacyl-tRNA complexes is the difficulty in preparing sufficient substrate in a form that is nonhydrolyzable under physiological conditions. Thus, to date, no structures of synthetases or editing domains bound to aminoacyl-tRNAs have been determined. Two crystal structures of EF-Tu in complex with GTP and two different aminoacyl-tRNAs have been elucidated because the ternary complex protects the aminoacyl-ester bond from hydrolysis (33, 34). The ribosome is a ribonucleoprotein (RNP) complex that also binds aminoacyl-tRNAs. Nonhydrolyzable ACCA-3′-NH-Pro conjugates have been used to probe the unique properties of the cyclic structure of Pro as a ribosome substrate (35). Future studies using nonhydrolyzable aminoacyl-tRNA acceptor stem analogs should provide additional insight into substrate selection by editing domains, as well as other enzymes and RNPs that recognize aminoacyl-tRNA substrates.
Another family of trans-editing enzymes that act on misacylated tRNAs is the d-aminoacyl-tRNA deacylases (DTD), which remove d-amino acids mischarged onto a variety of tRNAs (36–38). Recent NMR studies using the stable posttransfer substrate analogs d- and l-alanyl-3′-aminoadenosine indicated binding of the d-analog but not of the l-analog (32). The achiral glycyl-3′-aminoadenosine substrate analog also bound DTD. These data are consistent with the efficient deacylation of Gly-tRNAGly by all DTDs tested. Interestingly, these deacylases appear to reject substrates with l-chirality rather than specifically selecting substrates with d-chirality. Overexpression of DTD is toxic to Ec cells, and EF-Tu likely protects Gly-tRNAGly from normally low levels of DTD, as has been demonstrated in vitro (32). Similarly, we hypothesize that in the cell, EF-Tu protects Pro-tRNAPro from the low levels of ProXp-ala deacylation observed in our in vitro assays (Fig. 1), as we have previously shown for Ala-tRNAAla (6). DTDs are structurally homologous to the N-terminal editing domain of archaeal ThrRS, which deacylates (l-Ser-tRNAThr), and in both cases the 2′-OH of A76 has been shown to play a catalytic role, activating a water molecule for the deacylation reaction (32, 39, 40). Thus, as for the ProRS editing domain homologs, the role of the RNA and the lack of critical amino acid side chains within the active site suggest a primordial origin for these free-standing editing domains (9, 17).
Given the highly congruous nature of amino acids, efficient deacylation of mischarged RNAs has likely been a hallmark of life since the genesis of RNA adaptor-based aminoacylation (40). Many extant organisms use specialized trans-editing enzymes, which supplement the activity of the cis-editing domains present in nearly one-half of ARSs. Double- and triple-sieve size-based substrate discrimination are the primary biological mechanisms adopted by editing proteins. Such mechanisms are evolutionarily advantageous; subtle changes in the identity of active site residues can result in altered substrate specificity, allowing for rapid genetic adaptation to environmental stresses. Using a unique nonhydrolyzable substrate analog, we show that Cc ProXp-ala deviates from the canonical sieve-based paradigm, instead using a strategy that includes size-based exclusion and conformational and chemical selection. This multipronged approach takes advantage of the unique stereochemical features of Pro to ensure robust discrimination against the cognate amino acid while retaining efficient deacylation of Ala-tRNAPro.
Methods
Amino Acid Volume Calculations.
Each standard free amino acid (Ala, Cys, Gly, Pro, Ser, and Val) was obtained from the PyMOL library and used with no adjustments. Aze was built in PyMOL starting from proline; the Cδ carbon and its two hydrogens were removed, and a new bond was built between the Cγ carbon and the nitrogen. Abu was built in a similar manner from leucine; both Cδ carbons and their hydrogens were removed, and two additional hydrogens were added to the Cγ carbon. The newly built Aze and Abu were subjected to two-step energy minimization using the MMFF94 force field. Molecular volumes for each amino acid were calculated by loading each PDB file into the Volume Assessor module of the 3V webserver (3vee.molmovdb.org/volumeCalc.php) using a 1.5-Å probe radius and high grid resolution (0.5 Å voxel size) (41).
Preparation of Cc ProXp-ala.
All Cc ProXp-ala mutants were generated using the QuikChange Site-Directed Mutagenesis Kit (Agilent). WT and mutant Cc ProXp-ala variants were prepared essentially as described previously (6). In brief, the gene encoding Cc ProXp-ala was cloned into pET15b (Novagen). Protein expression from Ec BL-21-CodonPlus(DE3)-RIL (Agilent) was carried out by induction with 0.1 mM isopropyl β-d-1-thiogalactopyranoside (Gold Biotechnology) for 16–20 h at room temperature. Uniformly singly labeled ([U-15N]) and doubly labeled ([U-13C, 15N]) ProXp-ala samples were prepared in M9 minimal medium supplemented with 100 µM FeCl3, 1 µg/mL thiamine HCl, and 1 µg/mL biotin and containing 1 g/L [15N]-ammonium chloride (Cambridge Isotopes) as the sole nitrogen source or 3 g/L [13C]-glucose (Cambridge Isotopes) as the sole carbon source in addition to [15N]-ammonium chloride. His6-tagged ProXp-ala was purified on His-Select Nickel Affinity Gel (Sigma-Aldrich) using a 5–250 mM imidazole gradient. The His-tag was cleaved using the Thrombin Cleavage Capture Kit (Novagen) following the manufacturer’s suggested protocol. Cleaved His-tag and any remaining His-tagged proteins were captured by His-Select Nickel Affinity Gel (Sigma-Aldrich). Protein samples were further purified by size-exclusion chromatography (SEC) using a Superdex 200 Increase 10/300 column (GE Healthcare) and either NMR buffer (50 mM sodium phosphate pH 7.5 and 10 mM NaCl) or AUC buffer (50 mM Hepes pH 6.8, 30 mM KCl, and 1 mM MgCl2). Enzyme concentrations were determined using the Bio-Rad Protein Assay Kit.
Preparation of RNA.
Amino acids were obtained from Sigma-Aldrich, and [α-32P]-ATP was obtained from PerkinElmer Life Sciences. Ec tRNAPro was prepared by in vitro transcription using T7 RNA polymerase as described previously (42) and 3′-[32P]–labeled using tRNA nucleotidyltransferase as described previously (43). Preparation of aminoacylated Ec tRNAPro was carried out in 50 mM Hepes pH 7.5, 20 mM KCl, 4 mM ATP, 10 mM MgCl2, 0.1 mg/mL BSA, and 20 mM β-mercaptoethanol by incubating 10 µM editing-deficient K279A Ec ProRS, 10 µM tRNAPro, and 0.03 mg/mL pyrophosphatase (Roche) with 900 mM Ala, 30 mM Pro, 300 mM Aze, or 600 mM Abu for 10 min at 37 °C. Aminoacyl-tRNAs were phenol chloroform-extracted, followed by ethanol precipitation. Ser- and Cys-tRNAPro were prepared using dinitro-flexizyme and Ser-3,5-dinitrobenzyl ester (Ser-DBE) or Cys-DBE as described previously (44). Val-tRNAPro was prepared using enhanced flexizyme and Val-4-chlorobenzyl thioester (Val-CBT) (44). Substrates for deacylation assays were dissolved in 3 mM sodium acetate at pH 5.2 and stored at −80 °C before use.
Synthetic microhelixPro RNA used in NMR titrations and AUC experiments was purchased from GE Healthcare. The nh-Ala-microhelixPro containing a hydrolysis-resistant 3′-amide-linked Ala moiety (Fig. 2) was synthesized as described previously (45) using a solid-phase synthesis approach with 3′-aminoacylamino-3′-deoxyadenosine–modified functionalized solid supports (46) and 2′-O-TOM nucleoside phosphoramidites (47). Before the experiments, RNAs were refolded by heating at 80 °C for 2 min then 60 °C for 2 min, followed by the addition of MgCl2 to 10 mM and then slow cooling at room temperature and equilibration with either NMR or AUC buffer using SEC, as described above for protein preparation.
NMR Backbone Resonance Assignments and Chemical Shift Perturbation Analysis.
For NMR experiments, ProXp-ala samples in NMR buffer were supplemented with 10% D2O (vol/vol) as a lock signal and 0.001% 4,4-dimethyl-4-silapentane-1-sulfonic acid (DSS) (wt/vol) as an internal chemical shift reference. All spectra were recorded at 25 °C. Titration experiments were performed by recording HSQC spectra on [U-15N]-ProXp-ala on a Bruker DRX-800 spectrometer equipped with a TXI cryoprobe. Either microhelixPro or nh-Ala-microhelixPro was added to 0.1 mM ProXp-ala at 0.2, 0.4, 0.6, 0.8, 1, 1.5, and 2 molar equivalents. Sequential backbone assignments of the free proteins were obtained from triple-resonance NMR spectra: HNCO, HNCA, HNCOCA, HNCACB, and CBCA(CO)NH (29). NMR spectra were recorded on [U-13C/15N]-ProXp-ala (0.75 mM) on a Bruker DRX-600 spectrometer, also containing a TXI cryoprobe. Signals corresponding to backbone HN, N, CA, and CB atoms for 156 of the 160 (98%) non-Pro residues could be assigned. Backbone assignments of ProXp-ala in complex with microhelixPro were determined by the same method using 0.63 mM each of ProXp-ala and microhelixPro, resulting in assignment of 153 (96%) of the non-Pro residues. Backbone assignments of ProXp-ala in complex with nh-Ala-microhelixPro were inferred by monitoring CSPs during the titrations. Signals corresponding to 140 (88%) of the non-Pro residues could be assigned in this manner. Data were processed with NMRPipe (48) and analyzed with NMRViewJ (49). Chemical shifts were referenced directly (1H, 13C) or indirectly (15N) to DSS. Composite amide 1H and 15N CSPs were quantified as Δδ (ppm) = (ΔδH2 + ΔδN2/25)1/2. NMR resonance assignments have been deposited in the Biological Magnetic Resonance Data Bank, www.bmrb.wisc.edu (accession nos. 27185 and 27191).
Lineshape Analysis.
Dissociation constants and off-rates for NMR titrations were determined from 2D NMR lineshape analysis using TITAN software (https://www.ucl.ac.uk/biosciences/departments/smb/christodoulou/titan) (50). From spectra of the titration series described previously (eight points with 0–2 molar equivalents), 15 residues from Cc ProXp-ala titrations with microhelixPro and 14 residues from titrations with nh-Ala-microhelixPro were selected for fitting to a two-state ligand binding model, followed by bootstrap error analysis.
X-Ray Crystallography.
Cc ProXp-ala was crystallized via hanging drop vapor diffusion in 0.005 M trisodium citrate pH 5.6, 15% glycerol, and 25.5% wt/vol PEG4000 (Hampton Research). Diffraction datasets (collected at the NE-CAT beamlines, Advance Photon Source) were indexed, integrated, and scaled with the NE-CAT RAPD automated protocol (https://rapd.nec.aps.anl.gov/rapd). The phase was obtained by molecular replacement with PHASER, using the structure of Cc ProXp-ala (PDB ID code 1VJF) as a search model. Three molecules were found in the asymmetric unit and improved with iterative cycles of refinement with PHENIX (51). Crystallization and refinement statistics are detailed in Table S1, and the coordinates have been deposited into the PDB (PDB ID code 5VXB).
15N Relaxation Measurements.
Steady-state 1H-15N heteronuclear NOEs were obtained at 600 MHz at 25 °C using published pulse sequences (26–28). Samples contained 0.5 mM ProXp-ala, 0.5 mM ProXp-ala and microhelixPro, or 0.2 mM ProXp-ala and nh-Ala-microhelixPro in NMR buffer. The steady-state heteronuclear 1H-15N NOE values were obtained by recording spectra with and without a 1H presaturation period (9 s), in which all proton signals were saturated by a train of 90° pulses, applied before the start of the experiment. Intensity ratios were calculated using the hetNOE analysis tool within NMRViewJ (49). Owing to spectral overlap and/or lack of assignments, values could be determined for 136 of the 160 non-Pro residues of ProXp-ala, for 132 of 160 residues for ProXp-ala bound to microhelixPro, and for 130 of 160 residues for ProXp-ala bound to nh-Ala-microhelixPro.
15N CPMG Relaxation Dispersion.
Single quantum 15N CPMG relaxation dispersion experiments were carried out at 600 and 800 MHz at 25 °C using published pulse sequences (30) on a sample containing 0.7 mM ProXp-ala in NMR buffer. Spectra were recorded using a constant relaxation delay (Τrelax) of 40 ms, with CPMG frequencies (νCPMG) ranging from 0 to 1,500 Hz (25 points) at 800 MHz and from 0 to 1,250 Hz (23 points) at 600 MHz. Three repeated frequencies at both 800 MHz (50, 250, and 1,250 Hz) and 600 MHz (50, 300, and 900 Hz) were included for error analysis. Spectra were processed using NMRPipe (48), and peak intensities were extracted from the pseudo-3D datasets by fitting resonances to a Gaussian model using the nlinLS function. Dispersion curves were obtained for 119 residues, 14 of which exhibited significant exchange (Rex >2 s−1). GUARDD software was used to fit the data to the Carver–Richards equation describing two-state exchange (52). The nine residues located in helix α2 and in the β1-α2 loop of ProXp-ala were globally fit to obtain a single exchange rate (kex) and major state population (PA). The errors reported are estimated from 100 Monte Carlo simulations.
Deacylation Assays.
Aminoacyl-tRNAPro deacylation reactions were performed under single-turnover conditions at room temperature. Typical reactions contained 0.1 μM [32P]-labeled aminoacyl-tRNAPro and 0.75 μM Cc ProXp-ala in deacylation buffer (150 mM potassium phosphate pH 7.0, 5 mM MgCl2, and 0.1 mg/mL BSA). Reactions were initiated by mixing equal volumes of aminoacyl-tRNAPro and enzyme. At the indicated time points, 2-µL aliquots were quenched into a solution containing 2 units of P1 nuclease (Sigma-Aldrich) in 200 mM sodium acetate, pH 5.2. Product formation was monitored by separating aminoacyl-A76 from A76 on PEI-cellulose plates (EMD Millipore) using a mobile phase of 0.05% ammonium chloride/5% acetic acid. Radioactive products were detected by autoradiography using a Typhoon FLA 9500 (GE Healthcare) and quantified using ImageQuant TL 8 software (GE Healthcare). Observed rate constants (kobs) were obtained by fitting the time course for aminoacyl-tRNAPro deacylation with a single-exponential equation using SigmaPlot (Systat Software). Each reported rate constant is an average of three independent assays.
Inhibition assays were performed under steady-state conditions at room temperature in 50 mM Hepes pH 6.8, 30 mM KCl, 5 mM MgCl2, and 0.1 mg/mL BSA. Typical reactions contained 250 nM [32P]-labeled Ala-tRNAPro, 500 nM or 1 µM inhibitor (nh-Ala-microhelixPro, Pro-tRNAPro, or tRNAPro), and 20 µM ProXp-ala. At the indicated time points, reaction aliquots were quenched and analyzed as described above, except that the rate of deacylation was determined by fitting the data points with a linear equation.
Analytical Ultracentrifugation.
Analytical ultracentrifugation was performed using an Optima XL-I ultracentrifuge (Beckman Coulter) with an An50 Ti rotor and standard double-sector Epon centerpieces equipped with sapphire windows. Sedimentation velocity experiments were performed at 50,000 rpm at 20 °C in AUC buffer, monitoring absorbance at 260 nm. AUC data analyses were carried out using SEDFIT and SEDPHAT, available at http://sedfitsedphat.nibib.nih.gov (19, 20, 53).
For Kd determination, samples contained 1 μM microhelixPro or nh-Ala-microhelixPro and Cc ProXp-ala at concentrations ranging from 0.2 to 10 μM. For each sample, the sedimentation profiles were first fit to the Lamm equation to obtain the sedimentation coefficient distribution, c(s) (53). The c(s) distributions were integrated to create an isotherm of signal-weighted average sedimentation coefficients, sw, as a function of ProXp-ala concentration. Data were fit with mass action law models in SEDPHAT to obtain an estimated Kd, which served as a starting point for the global fit. The set of sedimentation profiles obtained for various ProXp-ala concentrations was then globally fit using the hetero-association model in SEDPHAT to obtain the Kd and sedimentation coefficient of the binary complexes. The sedimentation and extinction coefficients for unbound ProXp-ala and each RNA species were obtained from independent experiments and remained fixed during the global fit.
For analysis of ProXp-ala mutants, relative binding affinities were determined using a semiquantitative AUC-based assay. Sedimentation velocity experiments were performed as described above using 1.2 μM, 2.4 μM, 4.8 μM and 9.6 μM Cc ProXp-ala. The c(s) and sw values were obtained for each mutant, and the sw values were compared with those of 1.2 μM WT ProXp-ala to estimate relative binding affinities, as described in Fig. S1.
Computational Modeling.
The starting structure for docking simulations was prepared using the Ef ProRS crystal structure (PDB ID code 2J3L; residues 233–402) (22) as a template for homology modeling using SWISS-MODEL (54–56). This structure was used rather than the known crystal structure of Cc ProXp-ala (PBD ID code 1VJF) because the latter was crystallized with the α2 helix in the open rather than closed conformation. Semiflexible docking was performed using Autodock Vina (57) with the CCA-Ala ligand built in Avogadro (58). All rotatable bonds between nucleotides and within the amino acid were left unrestrained to allow for unbiased docking geometries. Based on homology with the INS domain of Ef ProRS and other INS-like domains (6), as well as CSP data, the side chains of the following active site residues were set to be flexible: Arg28, Val29, Lys45, Asn46, Leu47, Phe83, Gln86, Met89, Val100, and His130. Docking into a 25-Å cubic volume centered on the active site resulted in 2,000 successful docking poses, with ∼8% (162 poses) oriented such that the Ala moiety was in the active site and all atoms from the RNA were outside of the pocket as determined by visual inspection. Of these, a large cluster of 78 had a nearly identical position of the Ala atoms (heavy atom rmsd <1 Å), with the three best-scored docking poses also having nearly identical nucleotide positioning (heavy atom rmsd <1 Å). The top scoring pose was chosen as the representative for further analysis. Standard explicit solvent MD simulations were carried out using the AmberTools14 and AMBER14 program suites (59). The AMBER ff14SB force field was used for all nucleosides (60, 61). Constructs were first neutralized with sodium ions and solvated in an octahedral box of TIP3P water (62) with a 10.0-Å cutoff.
Minimization and equilibration were each conducted in two steps. During the first minimization, 2,000 total steps were performed, with 1,000 steps of steepest descent and a particle mesh Ewald (PME) implementation of constant volume periodic boundaries (63). The solute was held fixed using a positional restraint of 500 kcal/mol⋅Å2. In the second minimization step, 5,000 steps were performed, with 1,000 steps of steepest descent, and the solute was left unrestrained. The system was gradually heated from 0 K to 300 K in the first step of equilibration for a total of 100 ps at a time step of 2 fs while holding the solute fixed with a weak positional restraint of 10 kcal/mol⋅Å2. The PME implementation for a constant pressure periodic boundary was used for the second equilibration and production simulations. The SHAKE algorithm was used to constrain all bonds involving hydrogen (64), and Langevin dynamics was applied at a collision frequency of 1.0 ps−1 to control the temperature of the system. A second equilibration was used to relax the system for 900 ps under production conditions. Production simulations were performed in steps of 10 ns until the rmsd vs. time plot from the initial structure leveled off, indicating equilibrium. A further 40 ns of production simulation in steps of 10 ns was performed after equilibrium. All data analysis and statistical calculations were performed in the final 40 ns of each simulation. A random seed was used for all equilibration and production simulations to alleviate the possibility of synchronization artifacts of the simulations, which may result from the use of Langevin dynamics (65, 66). For all steps of simulation, a nonbonded cutoff of 8.0 Å was used. All equilibration and production simulations were performed using the GPU accelerated implementation of the PME MD in the sander module of Amber14 (67). PyMol (68) and VMD (69) were used for structure and trajectory visualization. Investigation of the active site conformations was done on snapshots taken at 30 ns, to ensure that the structures were stable for at least 10 ns before and after the selected timepoint.
Acknowledgments
We thank Drs. Alex Hansen and Chunhua Yuan of Ohio State University’s Campus Chemical Instrumentation Center for assistance with the NMR experimental setup and data processing. We also thank Irina Shulgina for assistance with protein preparation. This work was supported by National Institutes of Health Grants R01 GM113656 (to K.M.-F.) and T32 GM008512 (to E.M.D. and A.B.K.), US Department of Energy Grant GUP-51294 (to K.N.), Austrian Science Fund Grant FWF P27947 (to R.M.), Japan Society for the Promotion of Science (JSPS) Grant-in-Aid for Scientific Research (S) 26220204 (to H.S.), and JSPS KAKENHI Grant 15H05836 in Middle Molecular Strategy (to Y.G.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. S.A.M. is a guest editor invited by the Editorial Board.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 5VXB). NMR resonance assignments have been deposited in the BioMagResBank, www.bmrb.wisc.edu (accession nos. 27185 and 27191).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1703925114/-/DCSupplemental.
References
- 1.Mascarenhas A, Martinis SA, An S, Rosen AE, Musier-Forsyth K. Fidelity mechanisms of the aminoacyl-tRNA synthetases. In: Köhler C, Rajbhandary UL, editors. Protein Engineering. Springer; New York: 2009. pp. 153–200. [Google Scholar]
- 2.Ahel I, Korencic D, Ibba M, Söll D. Trans-editing of mischarged tRNAs. Proc Natl Acad Sci USA. 2003;100:15422–15427. doi: 10.1073/pnas.2136934100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.An S, Musier-Forsyth K. Trans-editing of Cys-tRNAPro by Haemophilus influenzae YbaK protein. J Biol Chem. 2004;279:42359–42362. doi: 10.1074/jbc.C400304200. [DOI] [PubMed] [Google Scholar]
- 4.Sokabe M, Okada A, Yao M, Nakashima T, Tanaka I. Molecular basis of alanine discrimination in editing site. Proc Natl Acad Sci USA. 2005;102:11669–11674. doi: 10.1073/pnas.0502119102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ruan LL, Zhou XL, Tan M, Wang ED. Human cytoplasmic ProX edits mischarged tRNAPro with amino acid, but not tRNA, specificity. Biochem J. 2013;450:243–252. doi: 10.1042/BJ20121493. [DOI] [PubMed] [Google Scholar]
- 6.Vargas-Rodriguez O, Musier-Forsyth K. Exclusive use of trans-editing domains prevents proline mistranslation. J Biol Chem. 2013;288:14391–14399. doi: 10.1074/jbc.M113.467795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fersht AR, Dingwall C. Establishing the misacylation/deacylation of the tRNA pathway for the editing mechanism of prokaryotic and eukaryotic valyl-tRNA synthetases. Biochemistry. 1979;18:1238–1245. doi: 10.1021/bi00574a019. [DOI] [PubMed] [Google Scholar]
- 8.Nureki O, et al. Enzyme structure with two catalytic sites for double-sieve selection of substrate. Science. 1998;280:578–582. doi: 10.1126/science.280.5363.578. [DOI] [PubMed] [Google Scholar]
- 9.Kumar S, Das M, Hadad CM, Musier-Forsyth K. Substrate specificity of bacterial prolyl-tRNA synthetase editing domain is controlled by a tunable hydrophobic pocket. J Biol Chem. 2012;287:3175–3184. doi: 10.1074/jbc.M111.313619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mursinna RS, Lincecum TL, Jr, Martinis SA. A conserved threonine within Escherichia coli leucyl-tRNA synthetase prevents hydrolytic editing of leucyl-tRNALeu. Biochemistry. 2001;40:5376–5381. doi: 10.1021/bi002915w. [DOI] [PubMed] [Google Scholar]
- 11.Hendrickson TL, et al. Mutational separation of two pathways for editing by a class I tRNA synthetase. Mol Cell. 2002;9:353–362. doi: 10.1016/s1097-2765(02)00449-5. [DOI] [PubMed] [Google Scholar]
- 12.Guo M, et al. Paradox of mistranslation of serine for alanine caused by AlaRS recognition dilemma. Nature. 2009;462:808–812. doi: 10.1038/nature08612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sokabe M, et al. The structure of alanyl-tRNA synthetase with editing domain. Proc Natl Acad Sci USA. 2009;106:11028–11033. doi: 10.1073/pnas.0904645106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.An S, Musier-Forsyth K. Cys-tRNA(Pro) editing by Haemophilus influenzae YbaK via a novel synthetase-YbaK-tRNA ternary complex. J Biol Chem. 2005;280:34465–34472. doi: 10.1074/jbc.M507550200. [DOI] [PubMed] [Google Scholar]
- 15.Ruan B, Söll D. The bacterial YbaK protein is a Cys-tRNAPro and Cys-tRNA Cys deacylase. J Biol Chem. 2005;280:25887–25891. doi: 10.1074/jbc.M502174200. [DOI] [PubMed] [Google Scholar]
- 16.Liu Z, et al. Homologous trans-editing factors with broad tRNA specificity prevent mistranslation caused by serine/threonine misactivation. Proc Natl Acad Sci USA. 2015;112:6027–6032. doi: 10.1073/pnas.1423664112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.So BR, et al. Substrate-mediated fidelity mechanism ensures accurate decoding of proline codons. J Biol Chem. 2011;286:31810–31820. doi: 10.1074/jbc.M111.232611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Das M, Vargas-Rodriguez O, Goto Y, Suga H, Musier-Forsyth K. Distinct tRNA recognition strategies used by a homologous family of editing domains prevent mistranslation. Nucleic Acids Res. 2014;42:3943–3953. doi: 10.1093/nar/gkt1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dam J, Velikovsky CA, Mariuzza RA, Urbanke C, Schuck P. Sedimentation velocity analysis of heterogeneous protein-protein interactions: Lamm equation modeling and sedimentation coefficient distributions c(s) Biophys J. 2005;89:619–634. doi: 10.1529/biophysj.105.059568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao H, Brautigam CA, Ghirlando R, Schuck P. Overview of current methods in sedimentation velocity and sedimentation equilibrium analytical ultracentrifugation. Curr Protoc Biol Sci. 2013;71:20.12:20.12.1–20.12.49. doi: 10.1002/0471140864.ps2012s71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shen Y, Bax A. Protein backbone and sidechain torsion angles predicted from NMR chemical shifts using artificial neural networks. J Biomol NMR. 2013;56:227–241. doi: 10.1007/s10858-013-9741-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Crepin T, Yaremchuk A, Tukalo M, Cusack S. Structures of two bacterial prolyl-tRNA synthetases with and without a cis-editing domain. Structure. 2006;14:1511–1525. doi: 10.1016/j.str.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 23.Wong FC, Beuning PJ, Nagan M, Shiba K, Musier-Forsyth K. Functional role of the prokaryotic proline-tRNA synthetase insertion domain in amino acid editing. Biochemistry. 2002;41:7108–7115. doi: 10.1021/bi012178j. [DOI] [PubMed] [Google Scholar]
- 24.Bartholow TG, et al. Strictly conserved lysine of prolyl-tRNA synthetase editing domain facilitates binding and positioning of misacylated tRNA(Pro) Biochemistry. 2014;53:1059–1068. doi: 10.1021/bi401279r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kumar S, Das M, Hadad CM, Musier-Forsyth K. Substrate and enzyme functional groups contribute to translational quality control by bacterial prolyl-tRNA synthetase. J Phys Chem B. 2012;116:6991–6999. doi: 10.1021/jp300845h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kay LE, Torchia DA, Bax A. Backbone dynamics of proteins as studied by 15N inverse detected heteronuclear NMR spectroscopy: Application to staphylococcal nuclease. Biochemistry. 1989;28:8972–8979. doi: 10.1021/bi00449a003. [DOI] [PubMed] [Google Scholar]
- 27.Barbato G, Ikura M, Kay LE, Pastor RW, Bax A. Backbone dynamics of calmodulin studied by 15N relaxation using inverse detected two-dimensional NMR spectroscopy: The central helix is flexible. Biochemistry. 1992;31:5269–5278. doi: 10.1021/bi00138a005. [DOI] [PubMed] [Google Scholar]
- 28.Grzesiek S, Bax A. The importance of not saturating H2O in protein NMR: Application to sensitivity enhancement and NOE measurements. J Am Chem Soc. 1993;115:12593–12594. [Google Scholar]
- 29.Cavanagh J, Fairbrother WJ, Palmer AG, Skelton NJ, Rance M. Protein NMR Spectroscopy: Principles and Practice. Elsevier Science; 2010. [Google Scholar]
- 30.Hansen DF, Vallurupalli P, Kay LE. An improved 15N relaxation dispersion experiment for the measurement of millisecond time-scale dynamics in proteins. J Phys Chem B. 2008;112:5898–5904. doi: 10.1021/jp074793o. [DOI] [PubMed] [Google Scholar]
- 31.Novoa EM, et al. Ancestral AlaX editing enzymes for control of genetic code fidelity are not tRNA-specific. J Biol Chem. 2015;290:10495–10503. doi: 10.1074/jbc.M115.640060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Routh SB, et al. Elongation factor Tu prevents misediting of Gly-tRNA(Gly) caused by the design behind the chiral proofreading site of D-aminoacyl-tRNA deacylase. PLoS Biol. 2016;14:e1002465. doi: 10.1371/journal.pbio.1002465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nissen P, et al. Crystal structure of the ternary complex of Phe-tRNAPhe, EF-Tu, and a GTP analog. Science. 1995;270:1464–1472. doi: 10.1126/science.270.5241.1464. [DOI] [PubMed] [Google Scholar]
- 34.Nissen P, Thirup S, Kjeldgaard M, Nyborg J. The crystal structure of Cys-tRNACys-EF-Tu-GDPNP reveals general and specific features in the ternary complex and in tRNA. Structure. 1999;7:143–156. doi: 10.1016/s0969-2126(99)80021-5. [DOI] [PubMed] [Google Scholar]
- 35.Melnikov S, et al. Molecular insights into protein synthesis with proline residues. EMBO Rep. 2016;17:1776–1784. doi: 10.15252/embr.201642943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Calendar R, Berg P. D-tyrosyl RNA: Formation, hydrolysis and utilization for protein synthesis. J Mol Biol. 1967;26:39–54. doi: 10.1016/0022-2836(67)90259-8. [DOI] [PubMed] [Google Scholar]
- 37.Soutourina J, Plateau P, Blanquet S. Metabolism of D-aminoacyl-tRNAs in Escherichia coli and Saccharomyces cerevisiae cells. J Biol Chem. 2000;275:32535–32542. doi: 10.1074/jbc.M005166200. [DOI] [PubMed] [Google Scholar]
- 38.Soutourina J, Plateau P, Delort F, Peirotes A, Blanquet S. Functional characterization of the D-Tyr-tRNATyr deacylase from Escherichia coli. J Biol Chem. 1999;274:19109–19114. doi: 10.1074/jbc.274.27.19109. [DOI] [PubMed] [Google Scholar]
- 39.Hussain T, Kamarthapu V, Kruparani SP, Deshmukh MV, Sankaranarayanan R. Mechanistic insights into cognate substrate discrimination during proofreading in translation. Proc Natl Acad Sci USA. 2010;107:22117–22121. doi: 10.1073/pnas.1014299107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hussain T, et al. Post-transfer editing mechanism of a D-aminoacyl-tRNA deacylase-like domain in threonyl-tRNA synthetase from archaea. EMBO J. 2006;25:4152–4162. doi: 10.1038/sj.emboj.7601278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Voss NR, Gerstein M. 3V: Cavity, channel and cleft volume calculator and extractor. Nucleic Acids Res. 2010;38(Web Server issue):W555–W562. doi: 10.1093/nar/gkq395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Beuning PJ, Musier-Forsyth K. Hydrolytic editing by a class II aminoacyl-tRNA synthetase. Proc Natl Acad Sci USA. 2000;97:8916–8920. doi: 10.1073/pnas.97.16.8916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ledoux S, Uhlenbeck OC. [3′-32P]-labeling tRNA with nucleotidyltransferase for assaying aminoacylation and peptide bond formation. Methods. 2008;44:74–80. doi: 10.1016/j.ymeth.2007.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Goto Y, Katoh T, Suga H. Flexizymes for genetic code reprogramming. Nat Protoc. 2011;6:779–790. doi: 10.1038/nprot.2011.331. [DOI] [PubMed] [Google Scholar]
- 45.Moroder H, et al. Non-hydrolyzable RNA-peptide conjugates: A powerful advance in the synthesis of mimics for 3′-peptidyl tRNA termini. Angew Chem Int Ed Engl. 2009;48:4056–4060. doi: 10.1002/anie.200900939. [DOI] [PubMed] [Google Scholar]
- 46.Steger J, Micura R. Functionalized polystyrene supports for solid-phase synthesis of glycyl-, alanyl-, and isoleucyl-RNA conjugates as hydrolysis-resistant mimics of peptidyl-tRNAs. Bioorg Med Chem. 2011;19:5167–5174. doi: 10.1016/j.bmc.2011.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Micura R. Small interfering RNAs and their chemical synthesis. Angew Chem Int Ed Engl. 2002;41:2265–2269. doi: 10.1002/1521-3773(20020703)41:13<2265::AID-ANIE2265>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 48.Delaglio F, et al. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 49.Johnson BA. Using NMRView to visualize and analyze the NMR spectra of macromolecules. Methods Mol Biol. 2004;278:313–352. doi: 10.1385/1-59259-809-9:313. [DOI] [PubMed] [Google Scholar]
- 50.Waudby CA, Ramos A, Cabrita LD, Christodoulou J. Two-dimensional NMR lineshape analysis. Sci Rep. 2016;6:24826. doi: 10.1038/srep24826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kleckner IR, Foster MP. GUARDD: User-friendly MATLAB software for rigorous analysis of CPMG RD NMR data. J Biomol NMR. 2012;52:11–22. doi: 10.1007/s10858-011-9589-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schuck P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and Lamm equation modeling. Biophys J. 2000;78:1606–1619. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: A web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 55.Biasini M, et al. SWISS-MODEL: Modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014;42:W252–258. doi: 10.1093/nar/gku340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bordoli L, et al. Protein structure homology modeling using SWISS-MODEL workspace. Nat Protoc. 2009;4:1–13. doi: 10.1038/nprot.2008.197. [DOI] [PubMed] [Google Scholar]
- 57.Trott O, Olson AJ. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hanwell MD, et al. Avogadro: An advanced semantic chemical editor, visualization, and analysis platform. J Cheminform. 2012;4:17. doi: 10.1186/1758-2946-4-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Case DA, et al. Amber 11. University of California; San Francisco: 2011. [Google Scholar]
- 60.Cornell WD, et al. A second-generation force field for the simulation of proteins, nucleic acids, and organic molecules. J Am Chem Soc. 1995;117:5179–5197. [Google Scholar]
- 61.Cheatham TE, 3rd, Young MA. Molecular dynamics simulation of nucleic acids: Successes, limitations, and promise. Biopolymers. 2000-2001;56:232–256. doi: 10.1002/1097-0282(2000)56:4<232::AID-BIP10037>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 62.Jorgensen WL, Chandrasekhar J, Madura JD. Comparison of simple potential functions for simulating liquid water. J Chem Phys. 1983;79:926–935. [Google Scholar]
- 63.York DM, Darden TA, Pedersen LG. The effect of long-range electrostatic interactions in simulations of macromolecular crystals: A comparison of the Ewald and truncated list methods. J Chem Phys. 1993;99:8345–8348. [Google Scholar]
- 64.Ryckaert JP, Ciccotti G, Berendsen HJC. Numerical integration of the Cartesian equations of motion of a system with constraints: Molecular dynamics of n-alkanes. J Comput Phys. 1977;23:327–341. [Google Scholar]
- 65.Uberuaga BP, Anghel M, Voter AF. Synchronization of trajectories in canonical molecular-dynamics simulations: Observation, explanation, and exploitation. J Chem Phys. 2004;120:6363–6374. doi: 10.1063/1.1667473. [DOI] [PubMed] [Google Scholar]
- 66.Sindhikara DJ, Kim S, Voter AF, Roitberg AE. Bad seeds sprout perilous dynamics: Stochastic thermostat-induced trajectory synchronization in biomolecules. J Chem Theory Comput. 2009;5:1624–1631. doi: 10.1021/ct800573m. [DOI] [PubMed] [Google Scholar]
- 67.Salomon-Ferrer R, Götz AW, Poole D, Le Grand S, Walker RC. Routine Microsecond molecular dynamics simulations with AMBER on GPUs, part 2: Explicit solvent particle mesh Ewald. J Chem Theory Comput. 2013;9:3878–3888. doi: 10.1021/ct400314y. [DOI] [PubMed] [Google Scholar]
- 68.Schrodinger LLC. The PyMOL Molecular Graphics System, Version 1.8. Schrodinger; New York: 2015. [Google Scholar]
- 69.Humphrey W, Dalke A, Schulten K. VMD: Visual molecular dynamics. J Mol Graph. 1996;14:33–38, 27-8. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]