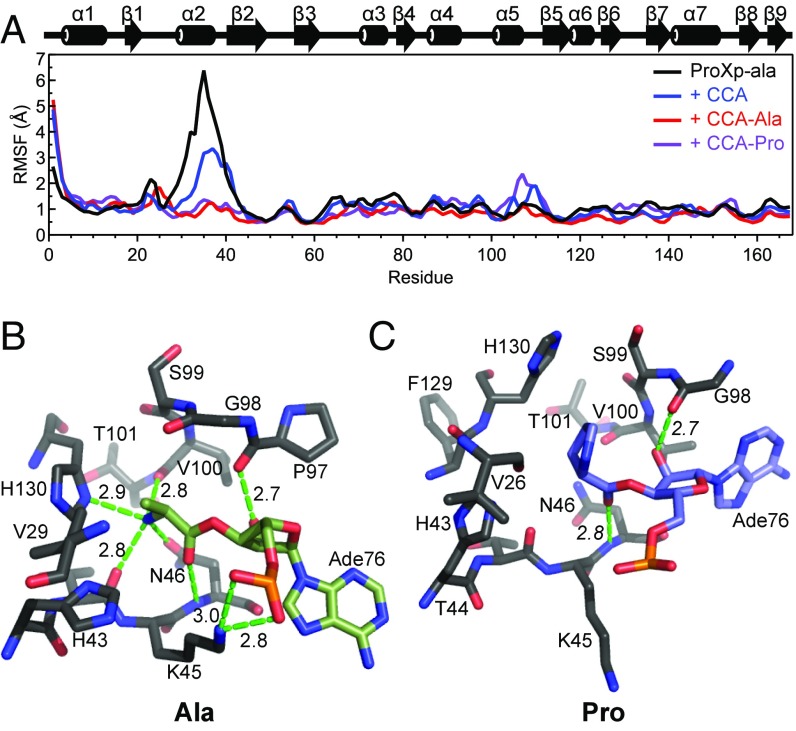
Fig. 6.
RMSFs and active site snapshots from MD simulations. (A) RMSF in backbone nitrogen atom coordinates for each residue of ProXp-ala either alone (black) or in the presence of a CCA trinucleotide (blue), CCA-Ala (red), or CCA-Pro (purple) over a 40-ns MD simulation trajectory. Secondary structure elements of ProXp-ala are displayed above the plot. (B and C) Substrate conformations in the Cc ProXp-ala active site after flexible docking and MD simulations for “mischarged” CCA-Ala (B, green) and cognate CCA-Pro (C, blue). A76-aa is shown in each case, and only ProXp-ala residues within at least 5 Å of the aminoacyl moiety are shown. Potential hydrogen bond distances are indicated in Å.