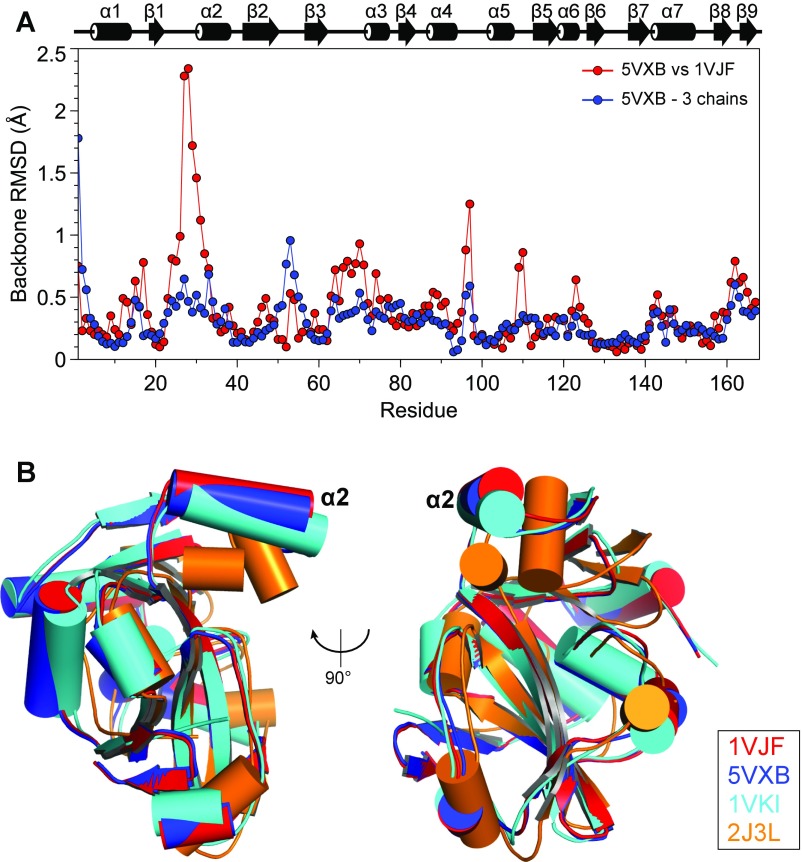
Fig. S5.
Comparison of the crystal structures of ProXp-ala and homologs. (A) Backbone RMSD values per residue between the two crystal structures of Cc ProXp-ala (PDB IDs 1VJF and 5VXB) and average backbone rmsd values per residue among the three monomers in the asymmetric unit of 5VXB. A secondary structure representation is displayed above the plot. (B) Superposition of Cα atoms of Cc ProXp-ala (red, PDB ID code 1VJF; blue, PDB ID code 5VXB), At ProXp-ala (cyan; PDB ID code 1VKI), and Ef ProRS INS (orange, PDB ID code 2J3L; residues 233–402). The alpha helix corresponding to Cc helix α2 is labeled.