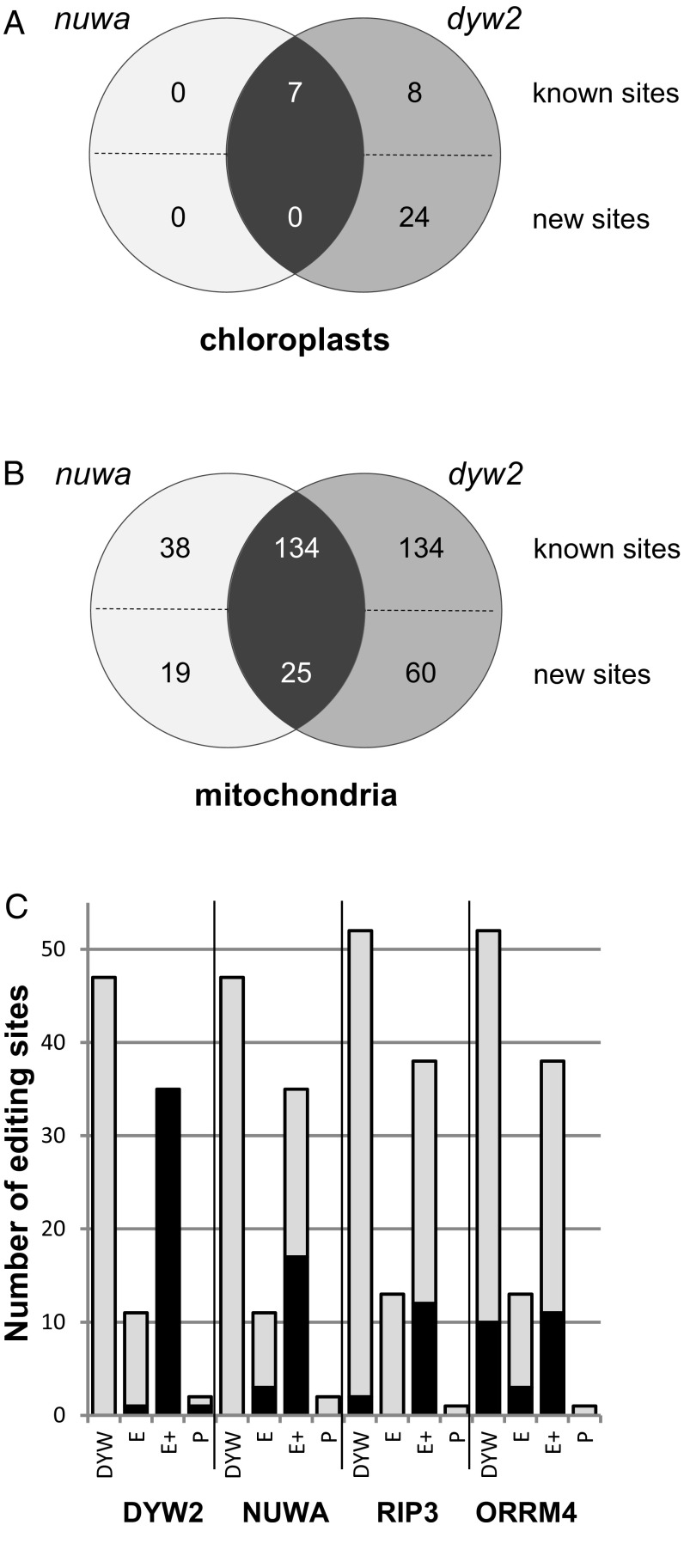
Fig. 3.
Editing activity of DYW2 and NUWA. The detailed results are provided in Dataset S2 A–D and Dataset S3. (A) Plastidial editing sites affected in dyw2 and nuwa. (B) Mitochondrial editing sites affected in dyw2 and nuwa. Venn diagrams summarizing the number of differentially edited sites in dyw2-1 and nuwa-2. The known sites correspond to the sites identified in Bentolila et al. (44), Sun et al. (24), Shi et al. (60), and Shi et al. (45). The new sites are the editing sites identified in this study. (C) DYW2 and NUWA-dependent sites are targeted by PPR-E+ proteins The PPR specificity of DYW2, NUWA, RIP3, and ORRM4 was estimated by counting the dependent (black) and independent (gray) editing sites associated with PPR of the various subfamilies [DYW, E, E+, and pure (P)]. An editing site is considered as depending on a particular protein if it is differentially edited between CT and mutant (P value < 5% after Bonferroni correction) and its editing extent is decreased by 10% or more in the mutant. It is independent otherwise. Editing sites associated with known PPRs are listed in Table S1 with their corresponding primary references. Values for RIP3 and ORRM4 were obtained by applying our statistical protocol to the raw data from Bentolila et al. (44) and Shi et al. (45), respectively. The total number of sites differs from one study to the other because of missing data from some editing sites in each study.