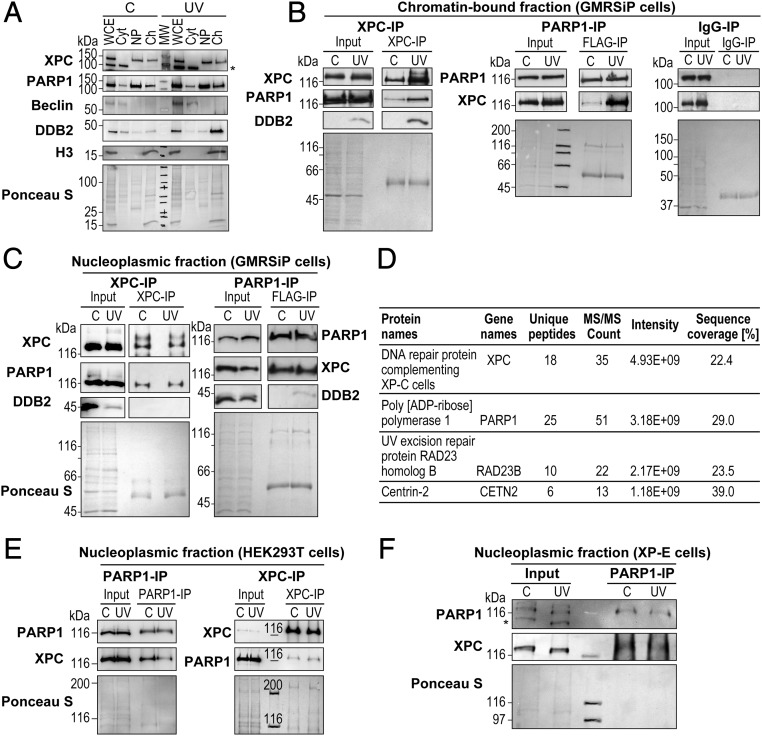
Fig. 1.
PARP1 interacts with XPC in the nucleoplasmic and chromatin fractions. (A) The GMRSiP cells expressing FLAG-tagged PARP1 were irradiated with 30 J/m2 UVC (or control), and whole-cell extracts (WCE) were fractionated to obtain cytoplasm (Cyt), nucleoplasm (Np), and chromatin-bound (Ch) fractions. The proteins from each fraction were immunoblotted for XPC, PARP1, and DDB2. Beclin and histone H3 were used as cytoplasmic and chromatin markers, respectively. The asterisk indicates a nonspecific band. The Ponceau S staining reflects the protein content at the end of each fractionation step. (B and C) The chromatin (B) and nucleoplasm fractions (C) of GMRSiP cells prepared as described above were subjected to IP for XPC, FLAG (PARP1), and mouse IgG (negative control), followed by the detection of PARP1, XPC, and DDB2. (D) Table showing the XPC-interacting proteins identified after XPC-IP of HEK293T cells followed by mass spectrometry analysis. (E) PARP1-IP and XPC-IP were performed in the Np fractions of control and UVC-treated HEK293T cells as shown in C. The Input and IP eluates were probed for XPC and PARP1. (F) PARP1-IP was performed in the Np fractions of control, and UVC-treated XP-E cells as described above. The Input and IP eluates were probed for XPC and PARP1. For B, C, E, and F the Ponceau S staining was used as loading control, and results shown here are representative of results from two to four experiments.