Fig. 4.
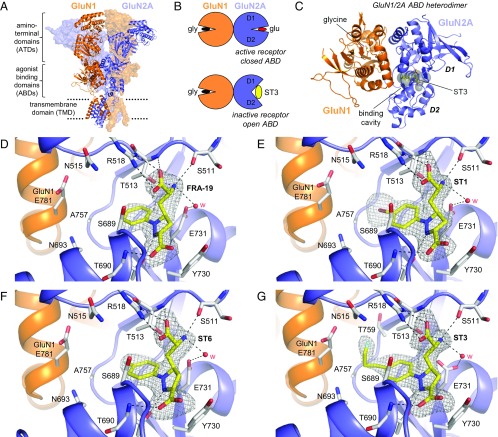
Structures of antagonist-bound GluN1/2A ABD heterodimers. (A) Structure of the NMDA receptor composed of two GluN1 and two GluN2 subunits (PDB ID code 4PE5) (3). Soluble NMDA receptor ABDs are expressed by deleting the ATD and replacing the transmembrane domain (TMD) with a dipeptide linker. (B) Cartoon illustrating the GluN1/2A ABD heterodimer with glycine bound in GluN1 and glutamate or ST3 bound in GluN2A. Binding of competitive antagonists stabilizes the cleft formed between the upper lobe (D1) and the lower lobe (D2) of the ABD in a more open conformation compared with binding of agonists. (C) Crystal structure of the GluN1/2A ABD heterodimer with bound glycine and ST3. The cavity (shown in gray) can be occupied by ACEPC ligands in the open ABD conformation and extends from the glutamate binding site toward the ABD dimer interface. The cavity was detected using CAVER (SI Materials and Methods). (D–G) Views of glutamate binding sites in GluN1/2A ABD heterodimer structures with bound FRA-19 (2.4 Å; D), ST1 (2.11 Å; E), ST6 (2.3 Å; F), and ST3 (1.95 Å; G). Select residues (gray carbon) within 4 Å of the ST3 ligand and potential polar interactions (dashed lines) are shown. Residue GluN2A H485 and backbone atoms for some residues are omitted for clarity. The antagonists could be unambiguously fitted to their electron densities shown as gray mesh (mFo − DFc omit map contoured at 3.0 σ; see Fig. S5 for contour level at 5.0 σ). See Table S3 for data collection and refinement statistics.