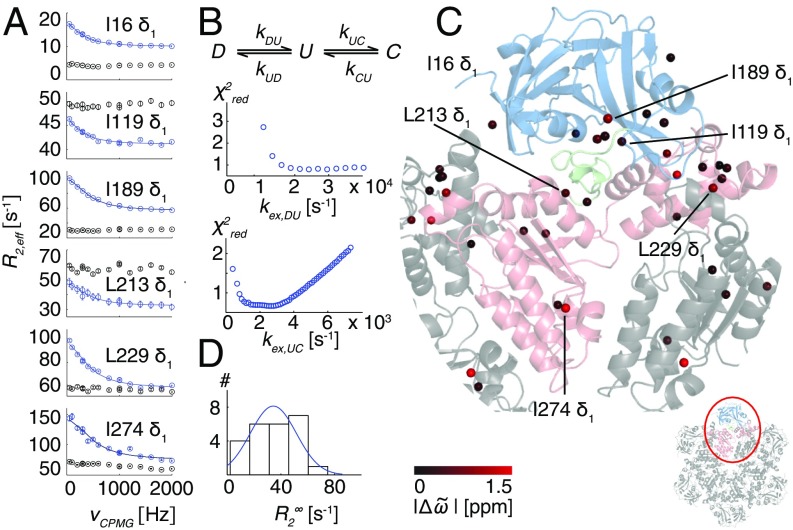
Fig. 2.
R95G p97-ADP disease mutant is plastic. (A) 13C-1H multiple quantum relaxation dispersion profiles (50 °C, 18.8T) for selected residues in the NTD (I16, I119), NTD–D1 linker (I189, L213), and the D1 domain (L229, I274) for R95G ND1Lp97-ADP (blue) and the corresponding molecule with the NTD domain covalently tethered to D1 (black). A global fit is included for the R95G profiles based on an analysis of 22 residues (Combined Fits of Chemical Shift and CPMG Data). (B) Three-state exchange model that takes into account both the 13C chemical shift data (Fig. 1B) and CPMG dispersion data that consists of the NTD up (U)/down (D) equilibrium and an additional process involving a third state, C. Reduced χ2 surfaces for rates for the first and second processes for the R95G mutant. (C) Positions of methyl groups from which CPMG dispersion profiles were analyzed via a global fit of the data using the model in B. Methyls are shown as spheres on the X-ray structure of ND1Lp97-ADP color-coded by the size of differences between 13C chemical shifts measured in spectra of R95G ND1Lp97-ADP and for the corresponding probes in state C (Combined Fits of Chemical Shift and CPMG Data), |Δϖ|. (D) Histogram of fitted rates for R95G ND1Lp97-ADP.