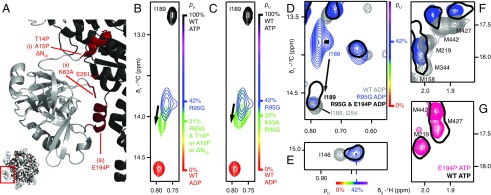
Fig. 3.
Destabilization of the NTD up state. (A) Three sites were targeted at the interface between the NTD and D1, as highlighted in red on the X-ray structure of WT ND1Lp97-ATPγS (29) and denoted by i–iii. (B) The N-terminal helix (site i) is destabilized through the substitution of a proline residue at positions 14 or 15 or removed by truncation, ΔN20. Superposition of small regions of 13C-1H HMQC spectra (18.8T, 50 °C) focusing on I189, with the I189δ1 cross-peak for ΔN20 R95G ND1Lp97-ADP in green (note that I189δ1 chemical shifts are identical for each of T14P, A15P, or ΔN20 substitutions), blue for R95G ND1Lp97-ADP, red for WT ND1Lp97-ADP, and black for WT ND1Lp97-ATP. Relevant mutations are placed in the R95G background because it is straightforward to evaluate changes to the up/down equilibrium in a system where pU ∼0.5. (C) As in B but for K63A,R95G ND1Lp97-ADP where an electrostatic interaction that stabilizes the up state has been removed (site ii). (D–F) Effect of removing the helix in the linker region connecting the NTD and D1 domains via the E194P mutation (site iii). The I189δ1 correlation in R95G,E194P ND1Lp97-ADP shifts (arrow) close to its position in WT ND1Lp97-ADP, indicative of an unfolding of the helix (D). Movement of the NTD into the down state does not occur as established by the positions of methyl correlations for I146 (E) and a number of methionines (F) that report on the up/down equilibrium that do not change with the E194P mutation (compare black single contour and blue contours in E and F). (G) Spectra of methionine methyls from WT ND1Lp97-ATPγS and E194P ND1Lp97-ATPγS superimpose, confirming that the helix in the NTD–D1 linker is not required for the NTD up state. pU values are highlighted on the sides of spectra in B–E, using the same color code as in Fig. 1.